- Research Services
- Capabilities
- About Us
- Resources
- Contact Us
 Microbiome and Colitis
Microbiome and Colitis Microbiome and Oncology
Microbiome and Oncology Microbiome and Other Disease Models
Microbiome and Other Disease Models Custom Models
Custom Models Microbiome Research
Microbiome ResearchBioModels offers customizable approaches to consider the microbiome in a variety of experimental systems. BioModels has the equipment, capabilities, and experience that is necessary to help you explore complex questions related to microbiome involvement in drug development and disease.
Assess your experimental microbiome-based test-articles for efficacy and mechanism of action with BioModels’ established approaches for modeling microbiome differences and perturbations in rodent models of colitis. The microbiome is an exciting and novel therapeutic target in Crohn’s Disease and Ulcerative Colitis. BioModels uses a number of approaches to model microbiome differences and perturbations for colitis projects, offering diverse experimental solutions such as:
Animal modeling with assessment of:
Assess your experimental microbiome-based test-articles for efficacy and/or mechanism of action with BioModels’ established approaches for modeling microbiome differences and perturbations in animal models of human cancer. The microbiome’s role in both cancer susceptibility and therapeutic response is increasingly appreciated, with particularly robust data supporting the microbiome’s impact on immune checkpoint inhibitor therapy efficacy.
BioModels has experience with a number of approaches to model microbiome differences and perturbations for oncology projects, offering diverse experimental solutions such as:
Animal modeling with:
Assess your experimental microbiome-based test-articles for efficacy and/or mechanism of action with BioModels’ established approaches for modeling microbiome differences and perturbations in animal models of disease. The microbiome is an exciting and novel therapeutic target in numerous disease states.
BioModels has used a number of approaches to model microbiome differences and perturbations for pre-clinical CRO projects, offering diverse experimental solutions with assessment of endpoints such as:
Animal modeling with:
The pace of drug discovery of rapidly increasing. BioModels collaborates frequently on proof-of-concept experiments designed to validate novel targets and approaches from the literature or your lab. While our internally optimized disease models are suitable for many projects, we understand that each program is unique, and it’s our goal to provide custom solutions to our clients. We have a history of success with custom modeling approaches for numerous disease states with microbiome components.
The BioModels team encourages detailed discussions and consultation aimed at identifying the most appropriate study designs based on our experience and the ever-evolving literature base. We are skilled in the replication of published data and routinely adapt literature-based methodologies to suit research needs. Some of these projects are more involved than others, but whatever stage your current project/program stands, we are happy to help and continue to push it forward.
Study Models

Differential response to DSS-induced colitis induction can be observed in animals sourced from different vendors, some of which are mediated by differences in their microbiome. Evaluation of colitis phenotypes in animals sourced from different vendors can be a straightforward approach to assess the microbiome’s impact in a given colitis model. Data from the acute DSS model induced in animals from different vendors is shown. When utilizing the DSS colitis model for assessment of novel therapeutics, we can help you select the most appropriate animal vendor to ensure the right microbiome for your experimental needs.
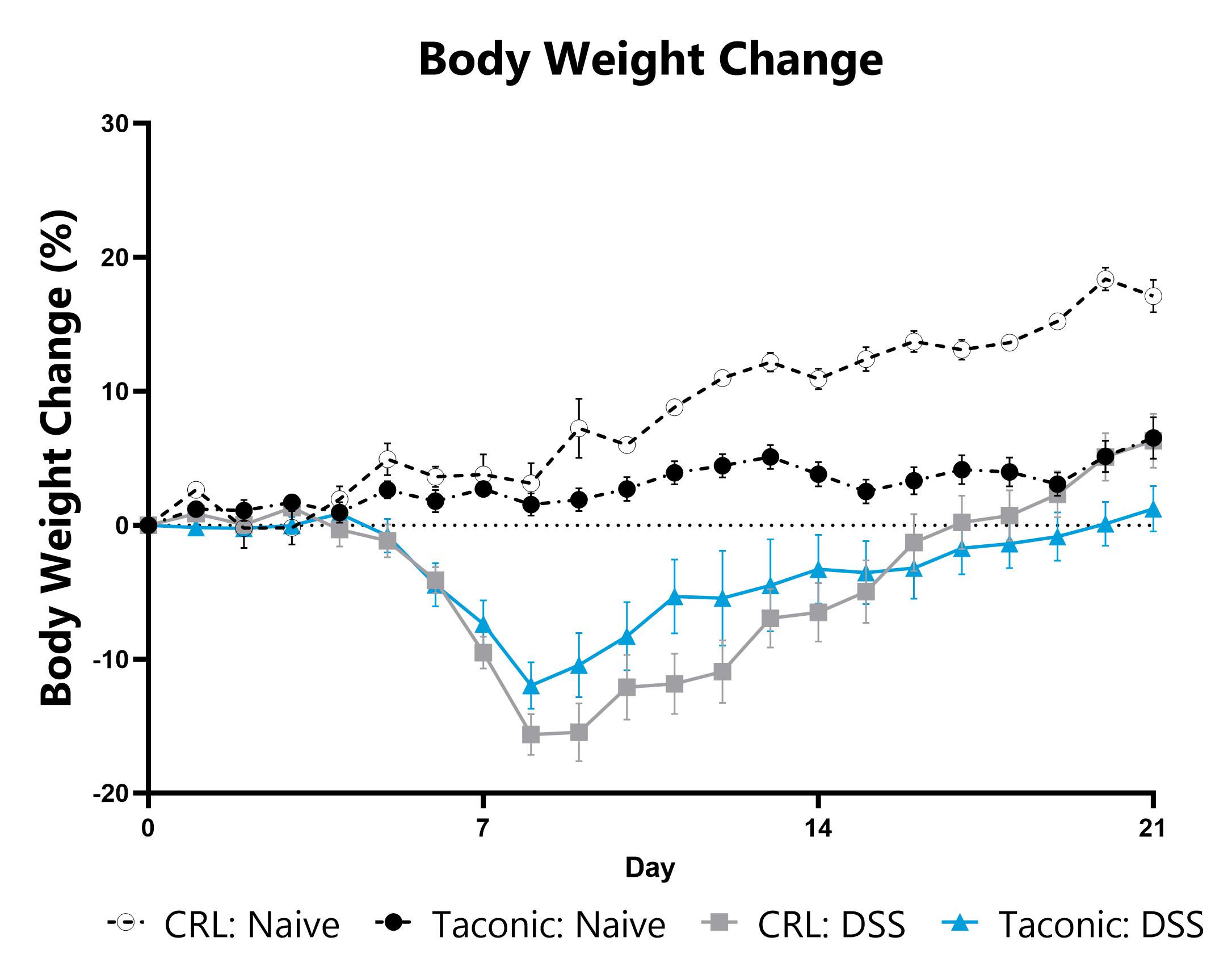
DSS-induced animals are weighed daily, and body weight change as compared to Day 0 is calculated.
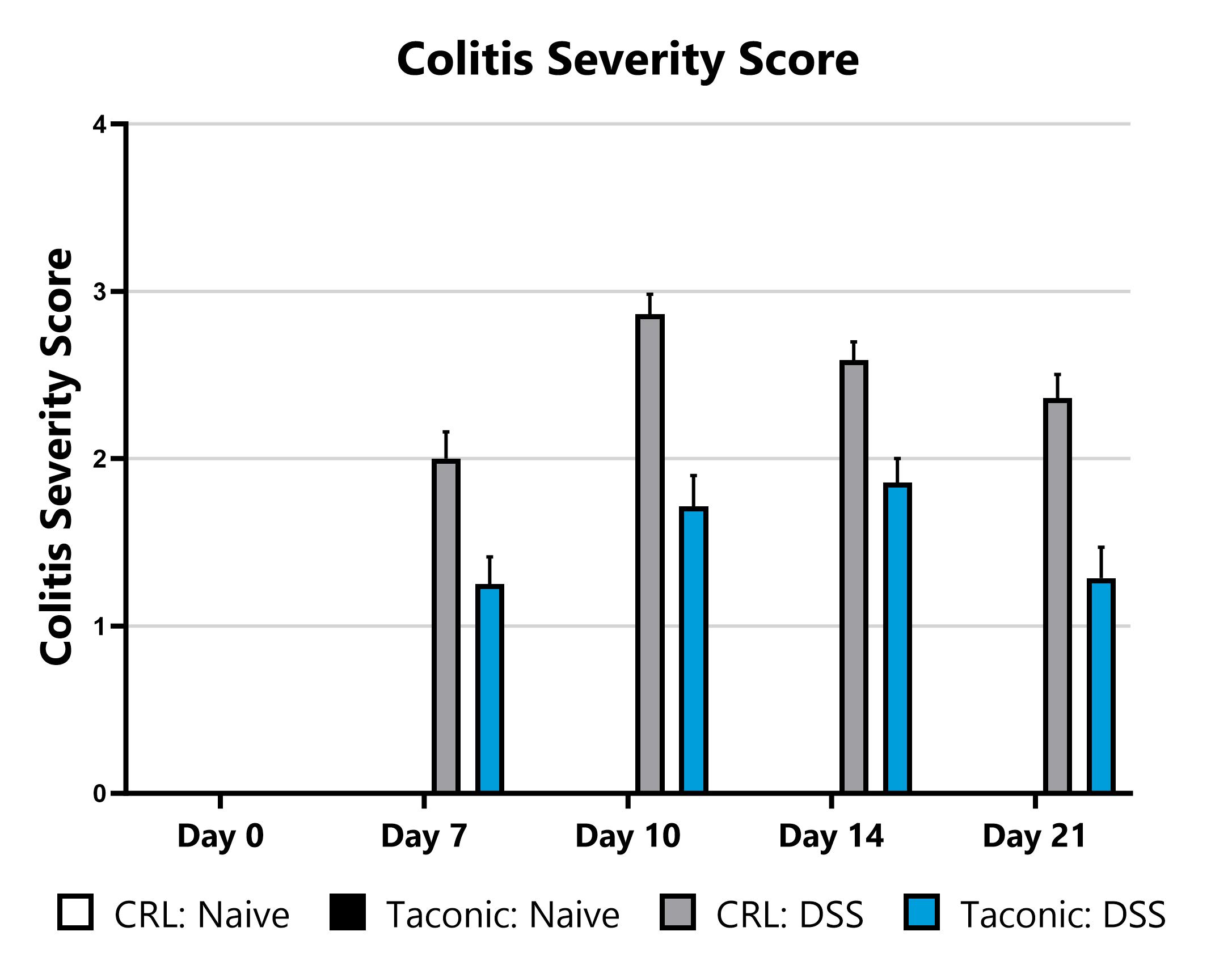
DSS-induced animals are assessed for colon inflammation by video endoscopy and are scored for colitis severity.

Close

Differential response to colitis induction can be observed in animals pre-treated with antibiotic, likely due to perturbation or reduction of the constitutive microbiome. Depending on the antibiotic pre-treatment paradigm selected, evaluation of colitis phenotype in antibiotic pre-treated animals enables assessment of phenotypes’ sensitivity toward broad microbiome depletion, species-specific depletion, or function-specific depletion. We can work with you to share our experience with antibiotic-induced microbiome modulation and how these treatments may impact your study goals. Data from the acute DSS model induced in animals pre-treated with an antibiotic cocktail intended for maximum microbiome depletion is shown.
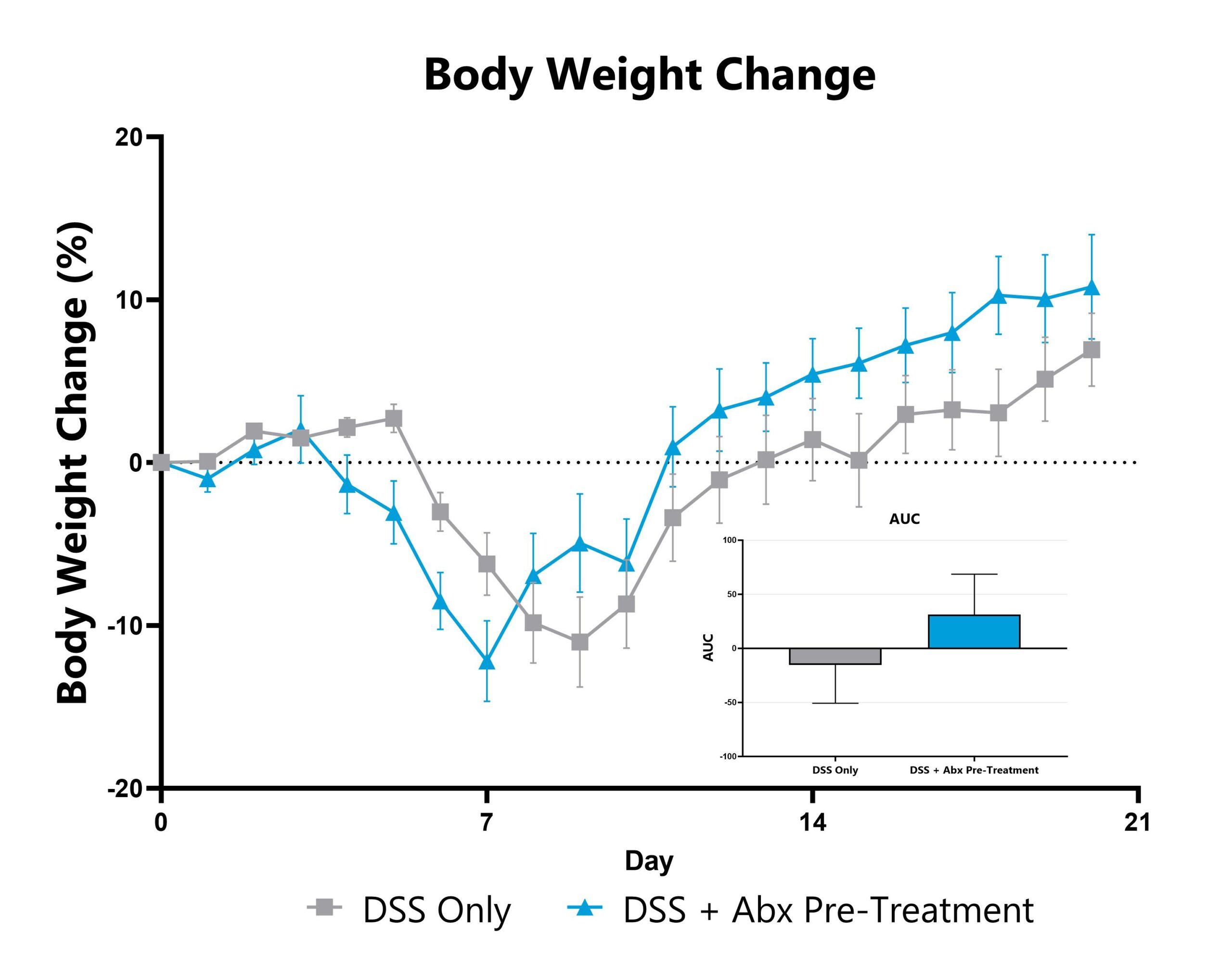
DSS-induced animals are weighed daily, and body weight change as compared to Day 0 is calculated. The AUC is calculated to compare treatment arms and is shown in the inset.
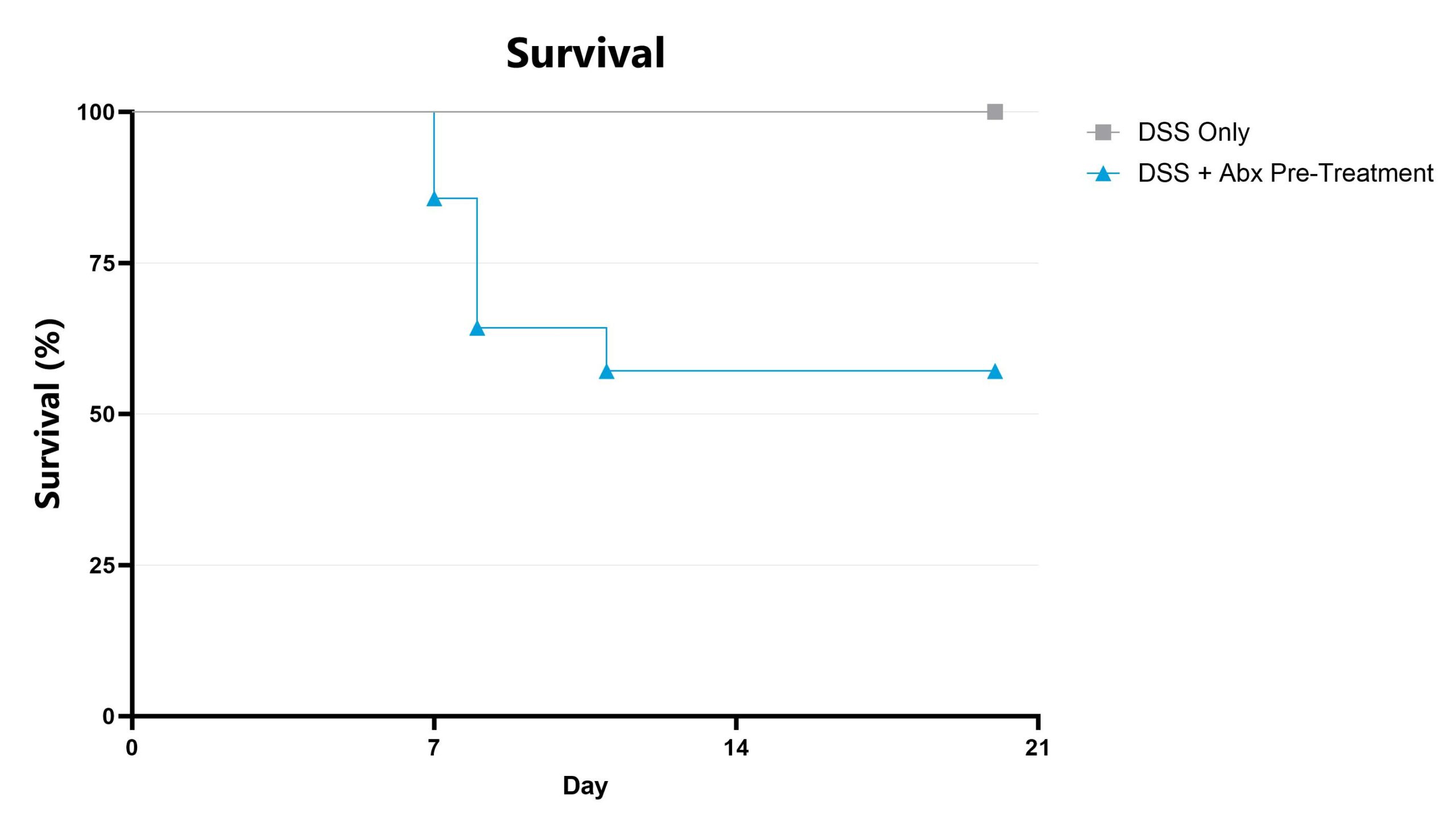
Animals are assessed for survival and moribundity on a daily basis.

Close

Differential response to colitis induction is observed in germ-free animals relative to those with an endogenous (normal) microbiome. Evaluation of colitis phenotypes in germ-free mice enables assessment of phenotypes’ dependency on a constitutive microbiome, whether due to the microbiome itself or due to other immunological effects. We can also use germ free animals to create custom microbiome colonized mice using your selected bacterial species or consortium. Data from the acute DSS model induced in germ-free mice is shown. Germ-free animals are housed in germ-free isolators, with germ-free husbandry practices as described in.
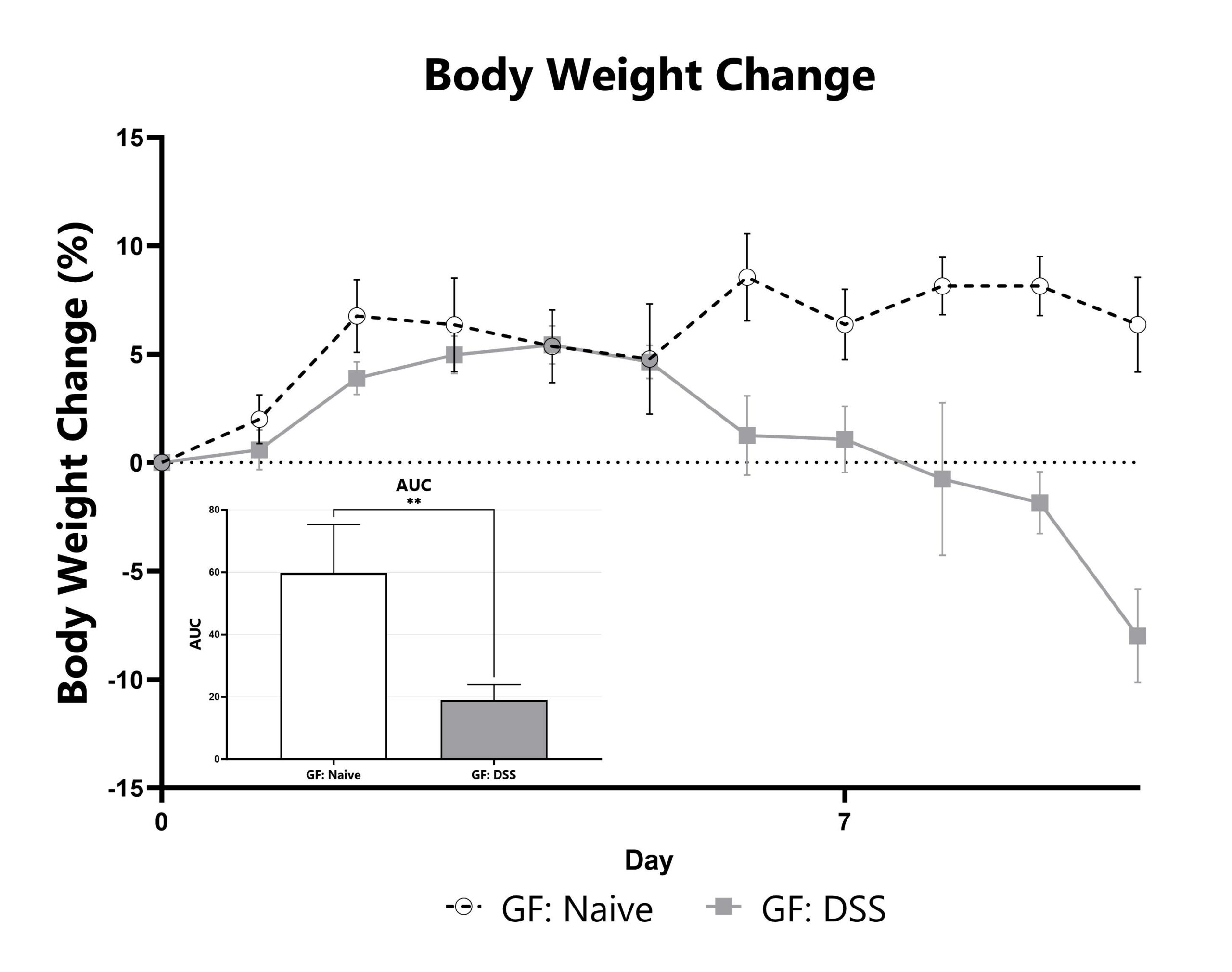
DSS-induced animals are weighed daily, and body weight change as compared to Day 0 is calculated. (**p<0.01)
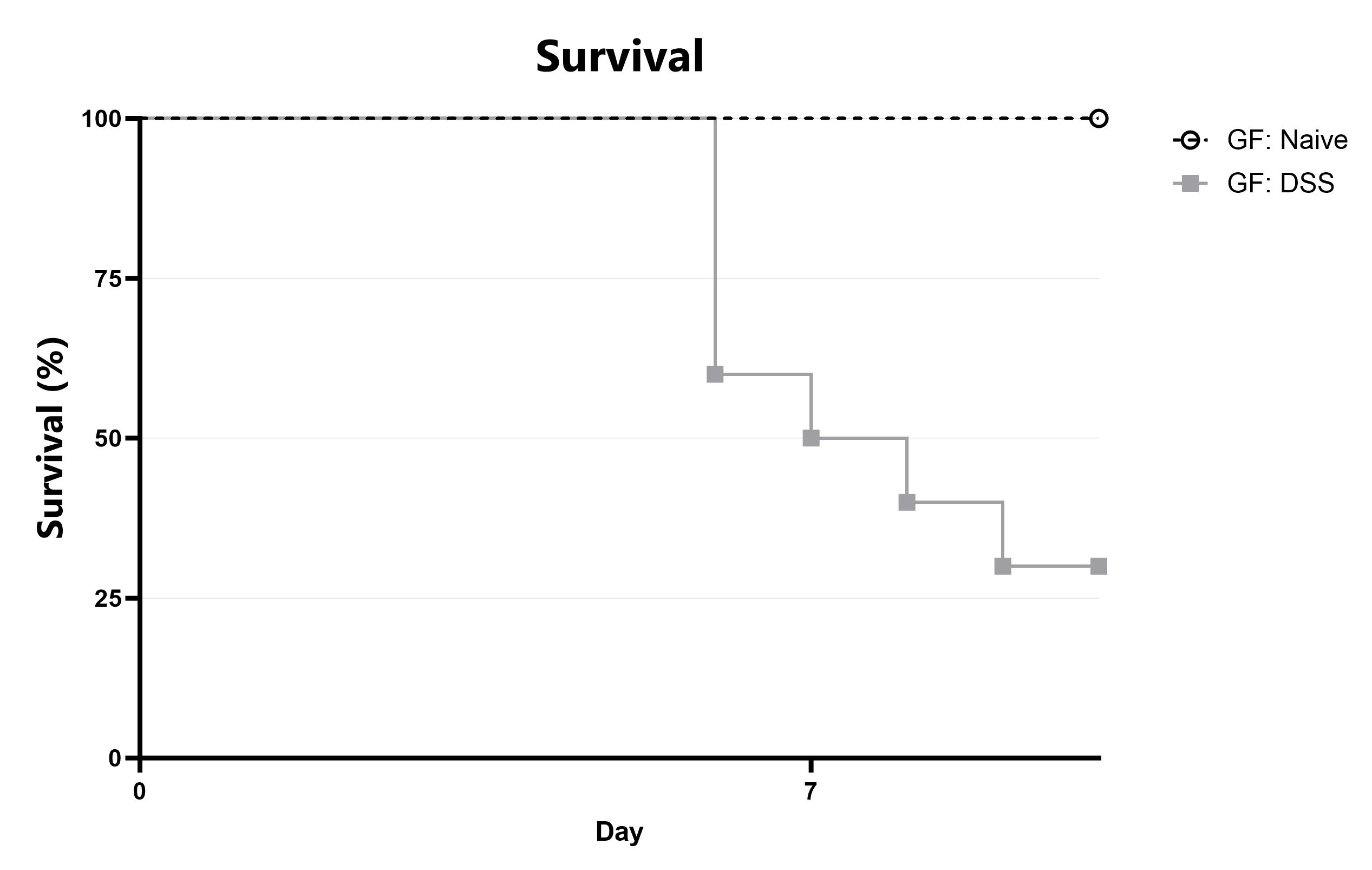
Animals are assessed for survival and moribundity on a daily basis.

Close

Differential response to TNBS-induced colitis induction can be observed in animals pre-treated with antibiotic, likely due to perturbation or reduction of the constitutive microbiome. Depending on the antibiotic pre-treatment paradigm selected, evaluation of colitis phenotype in antibiotic pre-treated animals enables assessment of phenotypes’ sensitivity toward broad microbiome depletion, species-specific depletion, or function-specific depletion. We can work with you to share our experience with antibiotic-induced microbiome modulation and how these treatments may impact your study goals.
Data from the acute TNBS model induced in animals pre-treated with an antibiotic cocktail intended for maximum microbiome depletion is shown.
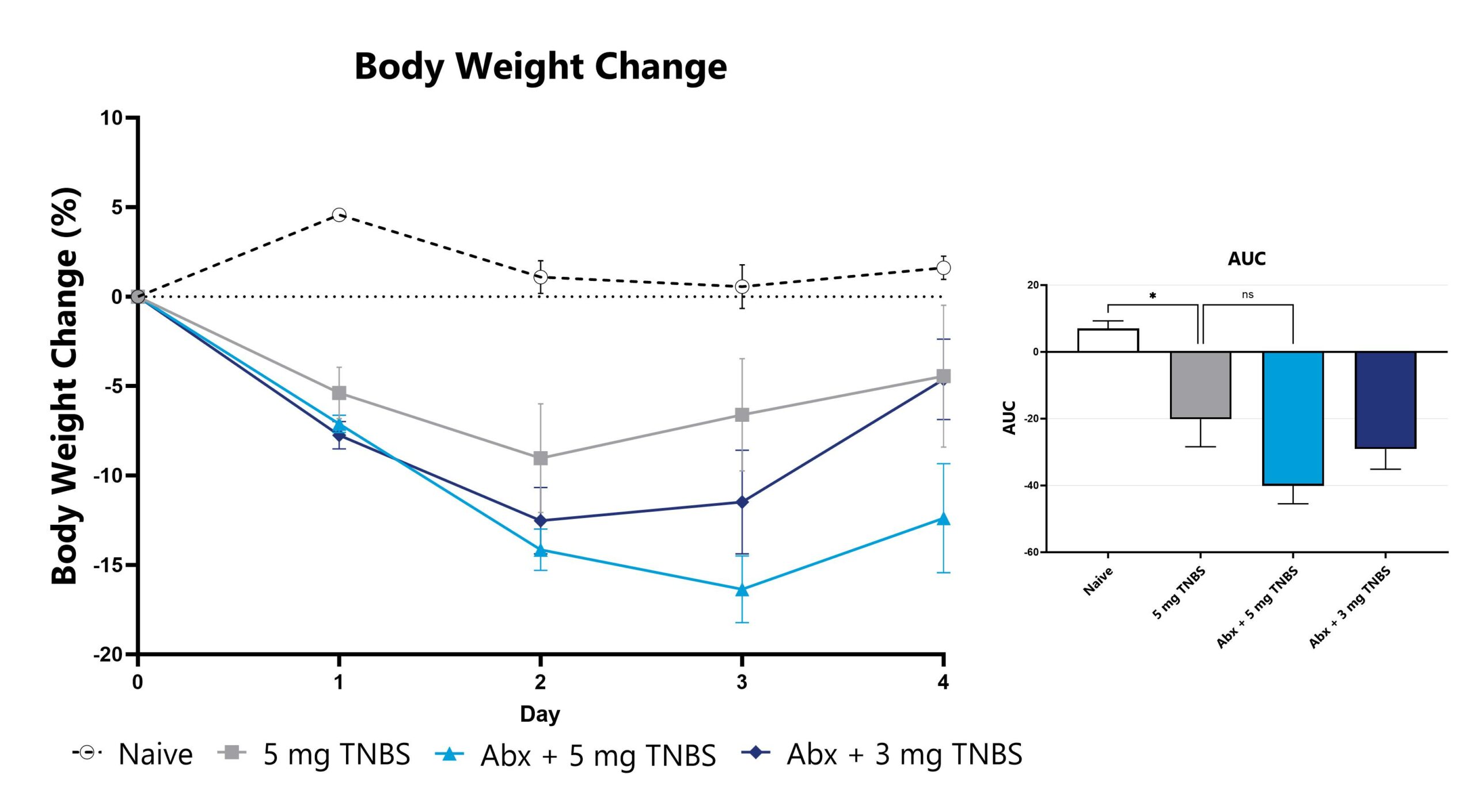
TNBS-induced animals are weighed daily, and body weight change as compared to Day 0 is calculated. (*p<0.05 as compared to vehicle control)
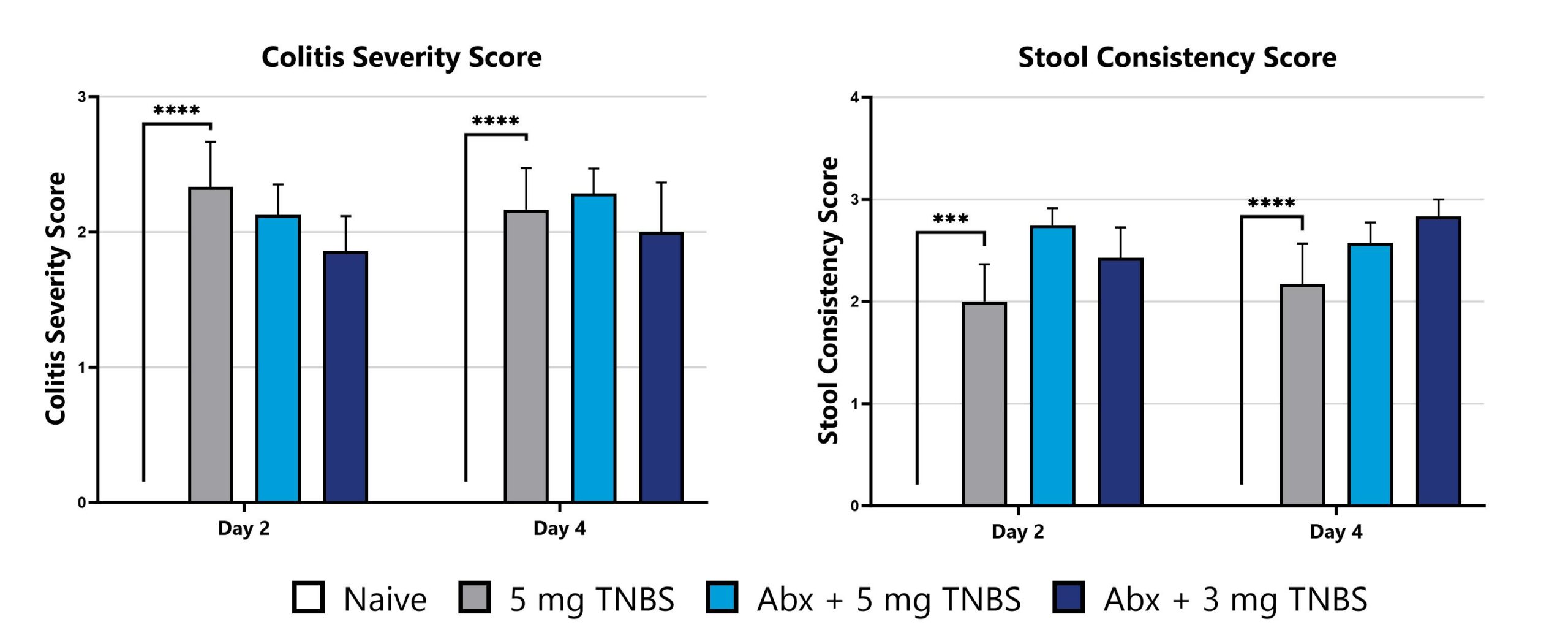
TNBS-induced animals are assessed for colon inflammation by video endoscopy and are scored for colitis severity and stool consistency. (***p<0.005; ****p<0.001 compared to the vehicle-control)

Close
Study Models

Differential responses in tumor growth can be observed in animals sourced from different vendors, some of which are mediated by vendor-differences in microbiome. Evaluation of tumor growth and checkpoint inhibitor sensitivity in animals sourced from different vendors can be a straightforward approach to assess the microbiome’s impact on these phenotypes. Data from a syngeneic subcutaneous melanoma model induced in animals from different vendors is shown.
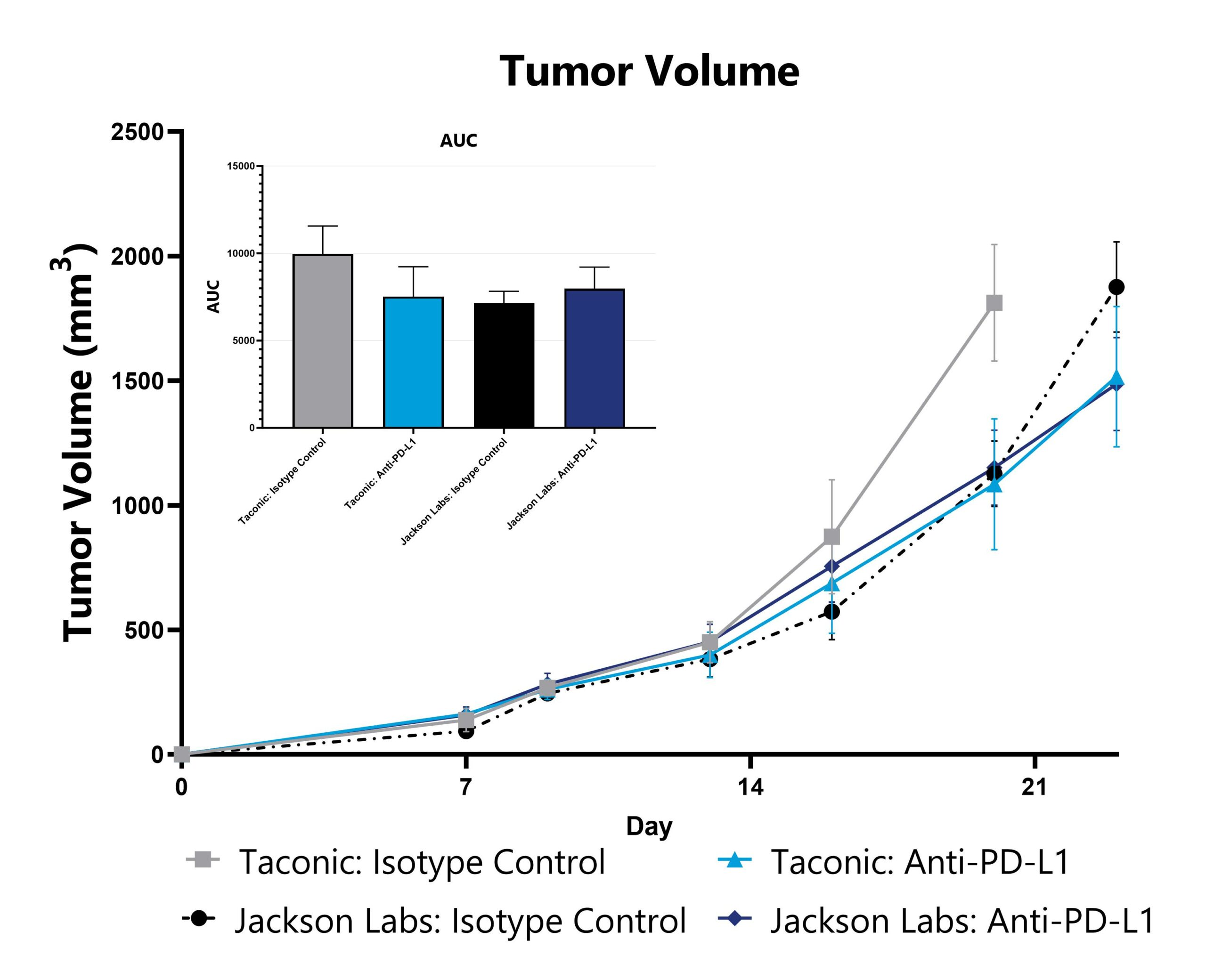
Tumor-inoculated animals undergo tumor measurement 2-3 times per week and tumor volume is calculated.

Close

Differential response to syngeneic tumor growth can be observed in germ-free animals relative to those with an endogenous microbiome. Evaluation of tumor growth and checkpoint inhibitor sensitivity in germ-free mice enables assessment of phenotypes’ dependency on a constitutive microbiome, whether due to the microbiome itself or due to other immunological effects. Data from a syngeneic subcutaneous melanoma model induced in germ-free mice is shown.
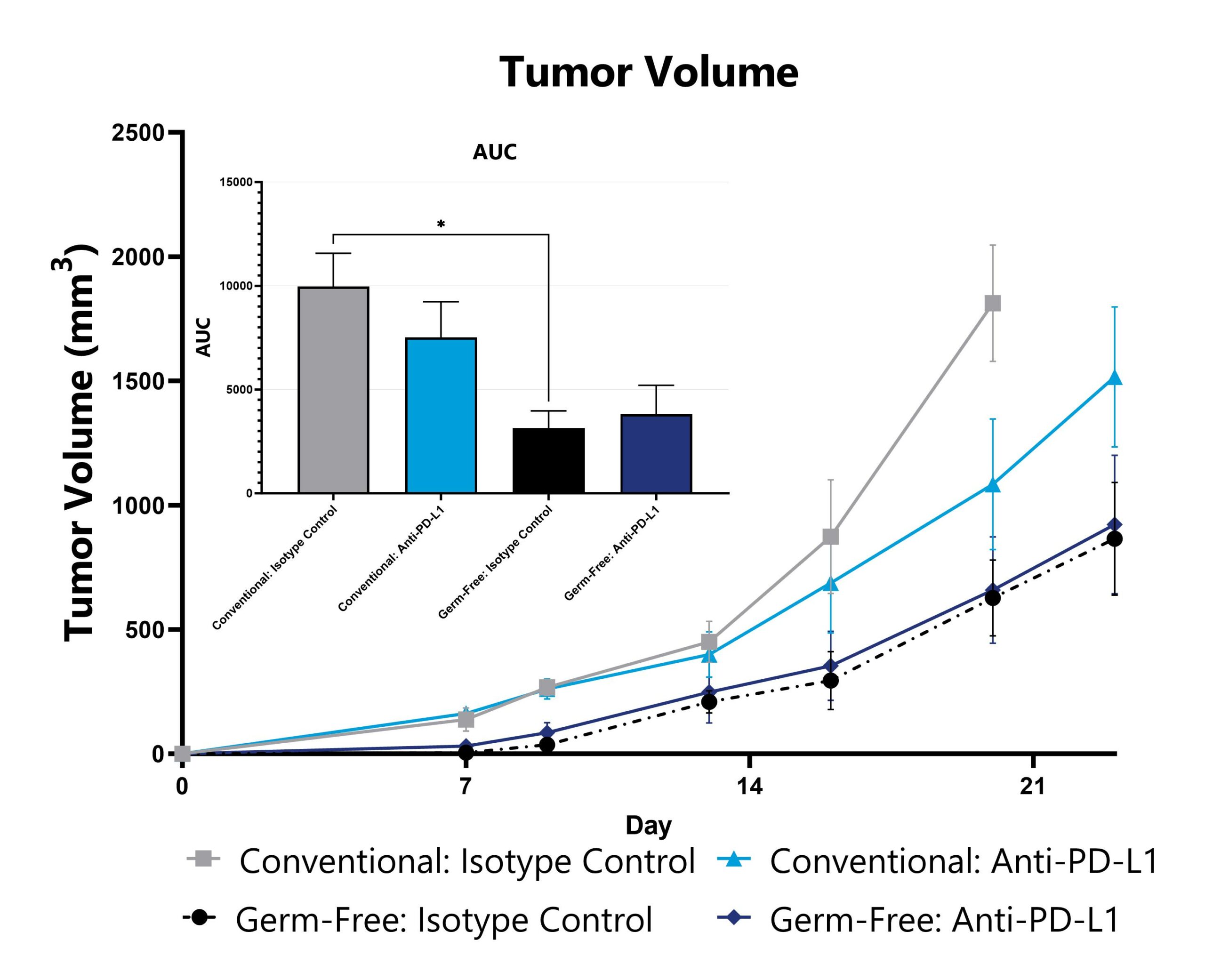
Tumor-inoculated animals undergo tumor measurement 2-3 times per week and tumor volume is calculated. The AUC is calculated to compare treatment arms and is shown in the inset. (*p<0.01).

Close
Study Models

Differential response to Graft versus Host Disease (GVHD) induction can be observed in animals sourced from different vendors, some of which are mediated by vendor-differences in microbiome. Evaluation of GVHD phenotypes in animals sourced from different vendors can be a straightforward approach to assess the microbiome’s impact in a given GVHD mode or with respect to your novel therapeutic approach. Data from the xenograft GVHD model induced in animals from different vendors is shown.
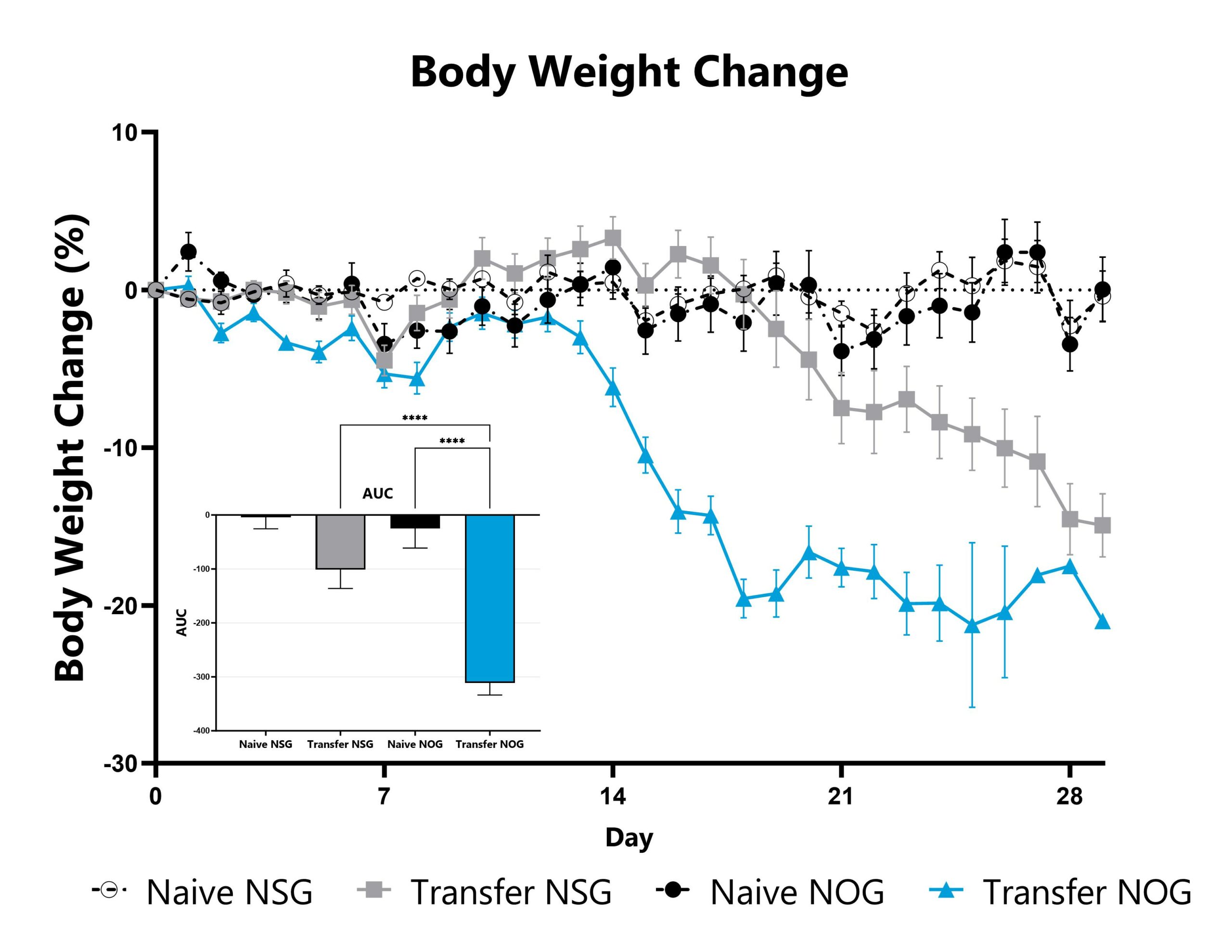
xGVHD animals are weighed daily, and body weight change as compared to Day 0 is calculated. The AUC is calculated to compare treatment arms and is shown in the inset. (****p<0.001).
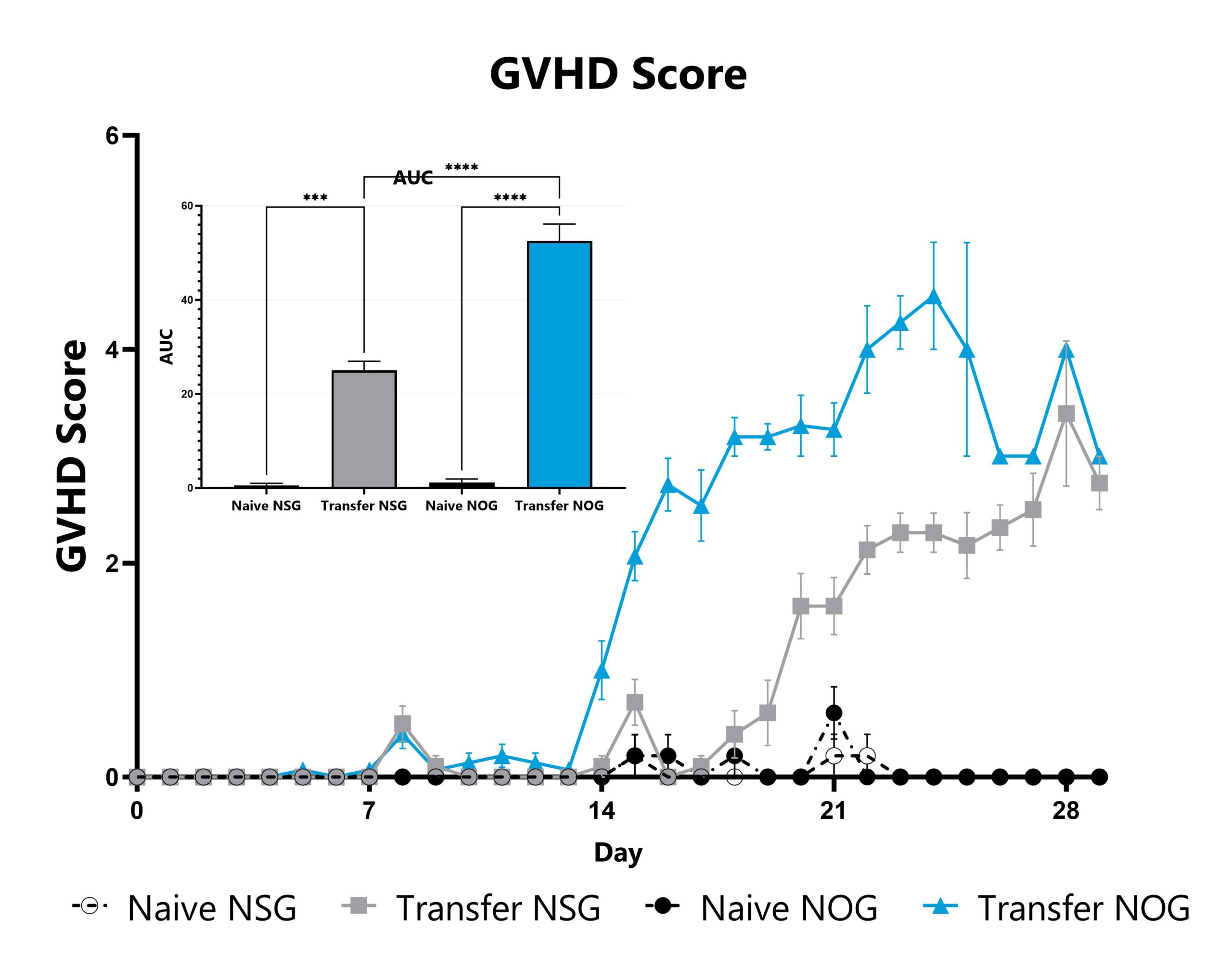
xGVHD animals are evaluated daily for weight loss, posture, activity, fur texture, and skin integrity phenotypes. Each phenotype is scored and used calculate a GVHD score. The AUC is calculated to compare groups and is shown in the inset. (***p<0.005; ****p<0.001).
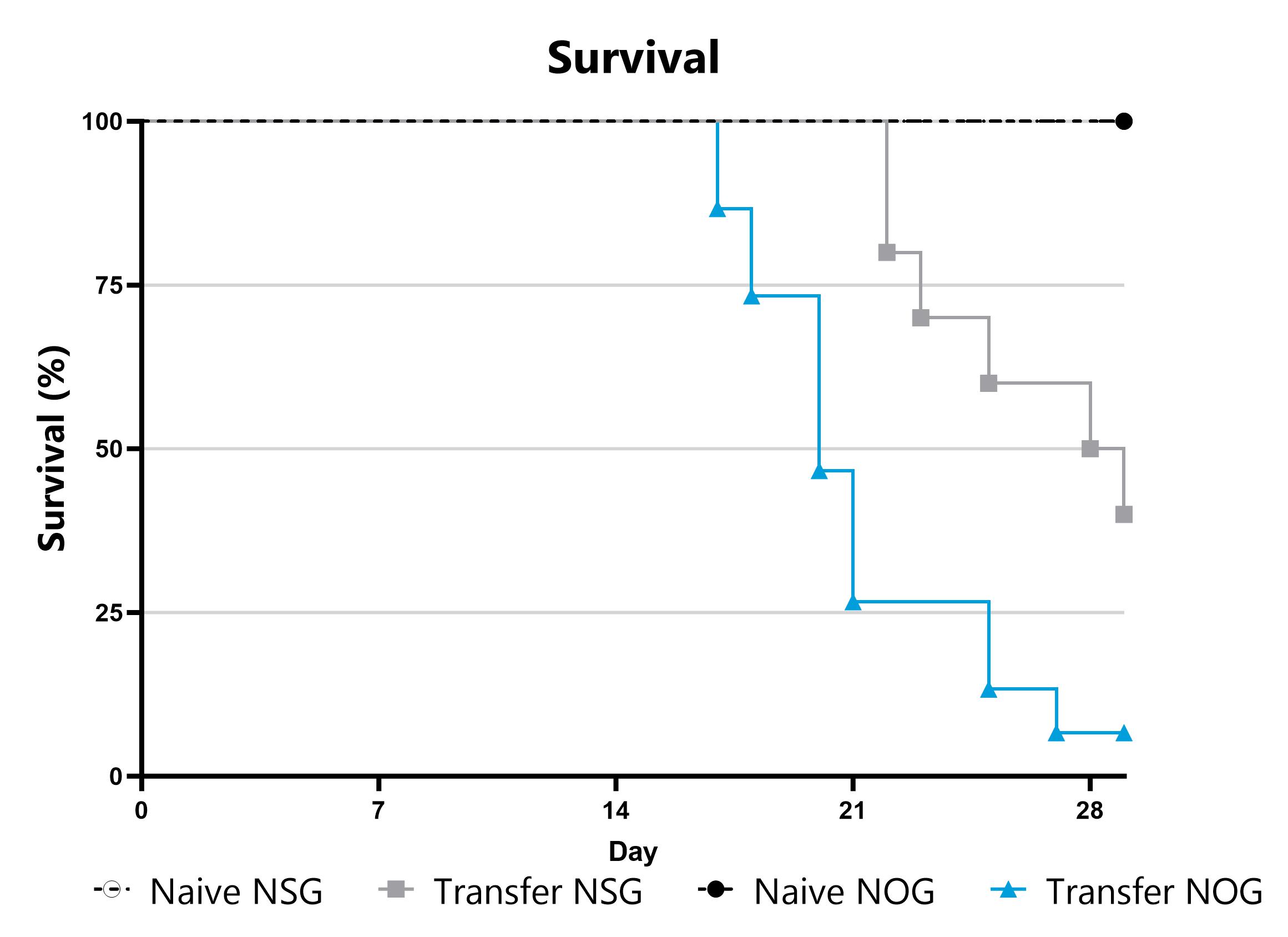
Animals are evaluated for survival and moribundity on a daily basis.
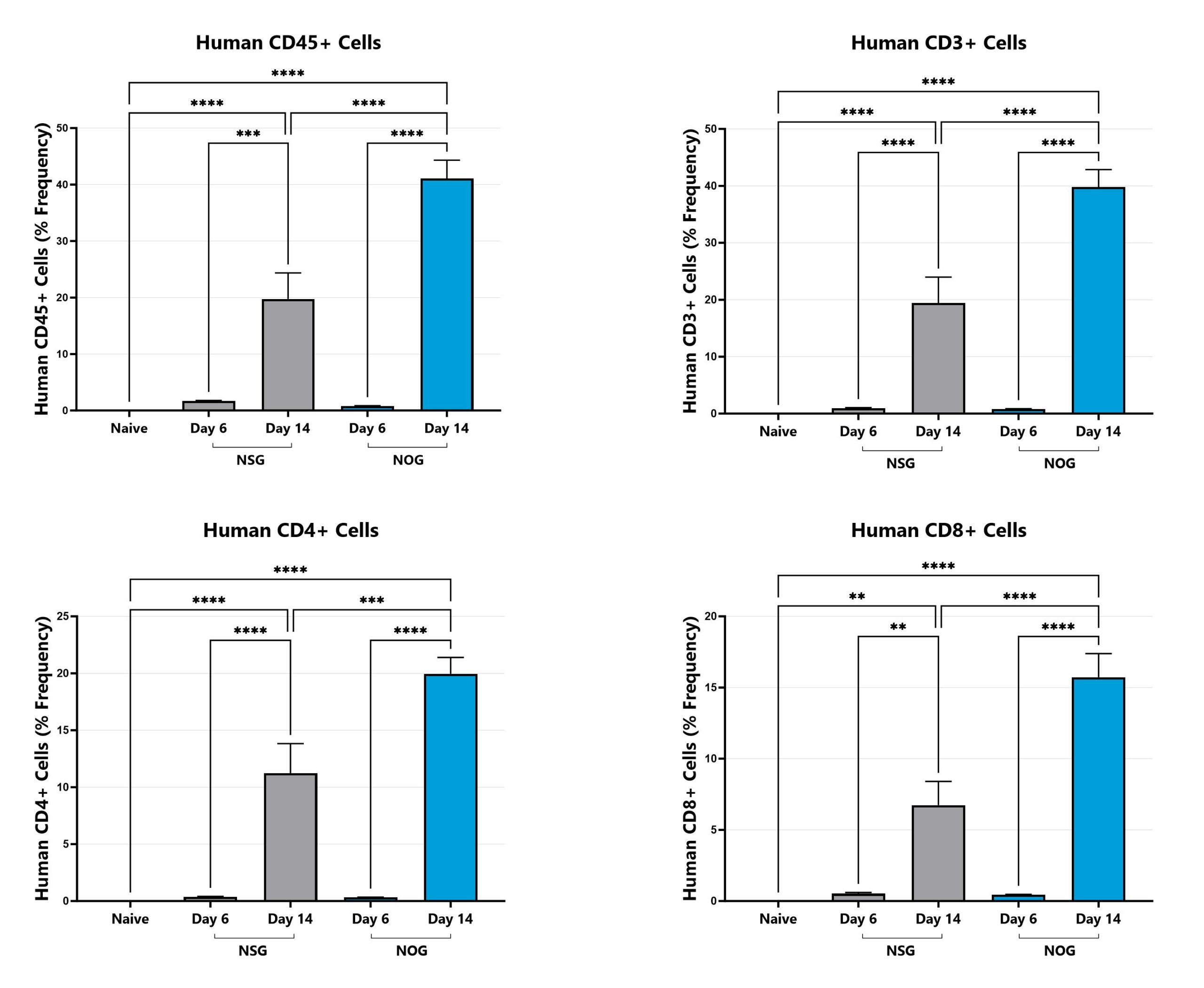
Animals are monitored for human leukocyte engraftment via flow cytometry over time. (**p<0.01; ***p<0.005; ****p<0.001).

Close

Differential responses to metabolic disease can be observed in animals pre-treated with antibiotic, likely due to perturbation or reduction of the constitutive microbiome. Depending on the antibiotic pre-treatment paradigm selected, evaluation of obesity and metabolism phenotype in antibiotic pre-treated animals enables assessment of phenotypes’ sensitivity toward broad microbiome depletion, species-specific depletion, or function-specific depletion. Data assessing metabolic phenotypes in normal mice, as well as from the high fat diet-induced obesity model, utilizing animals pre-treated with an antibiotic cocktail intended for maximum microbiome depletion is shown. A subset of animals were treated with a murine fecal microbial transplant (FMT).
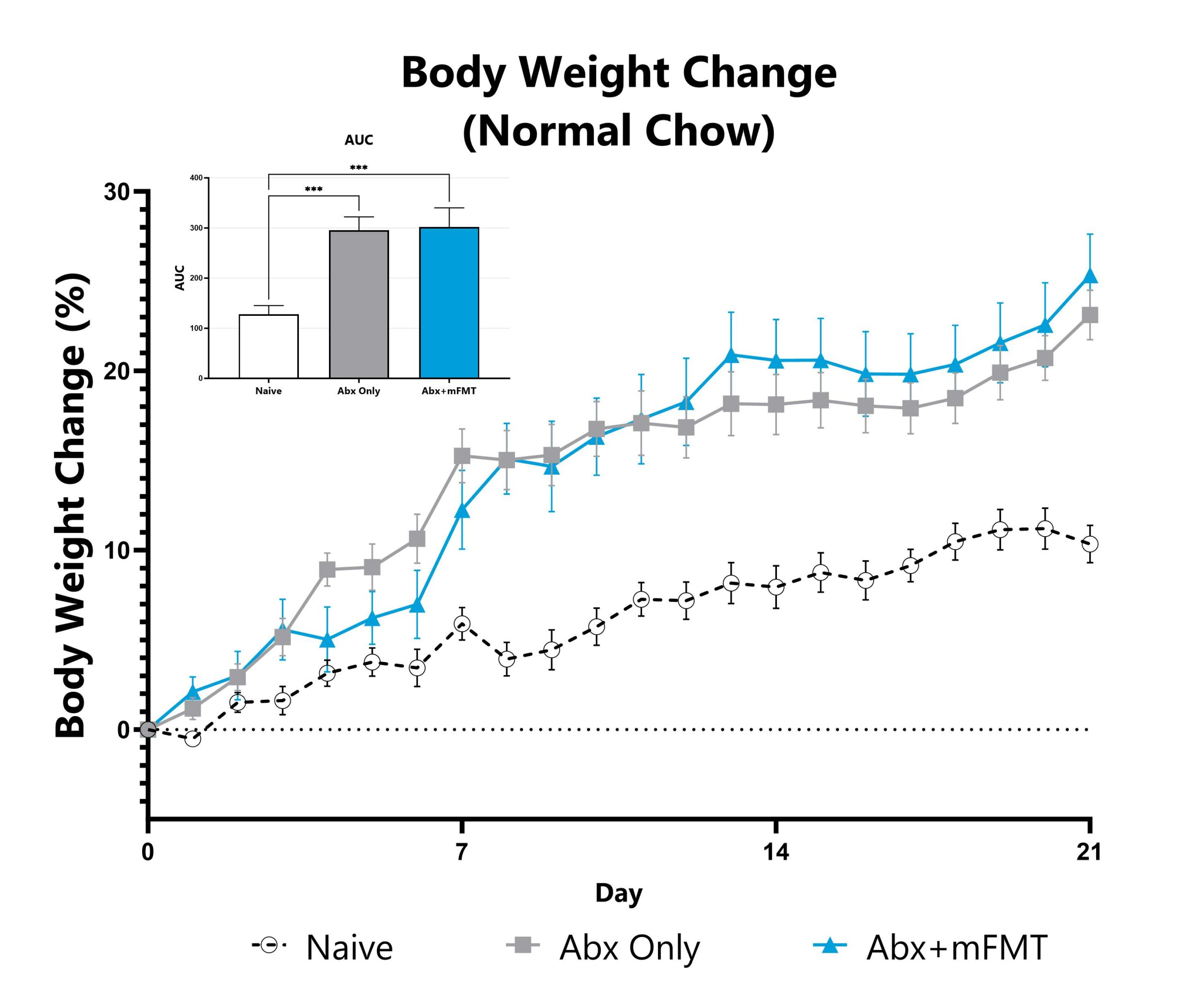
Animals are weighed daily, and body weight change as compared to Day 0 is calculated. The AUC is calculated to compare treatment arms and is shown in the inset. (***p<0.005).
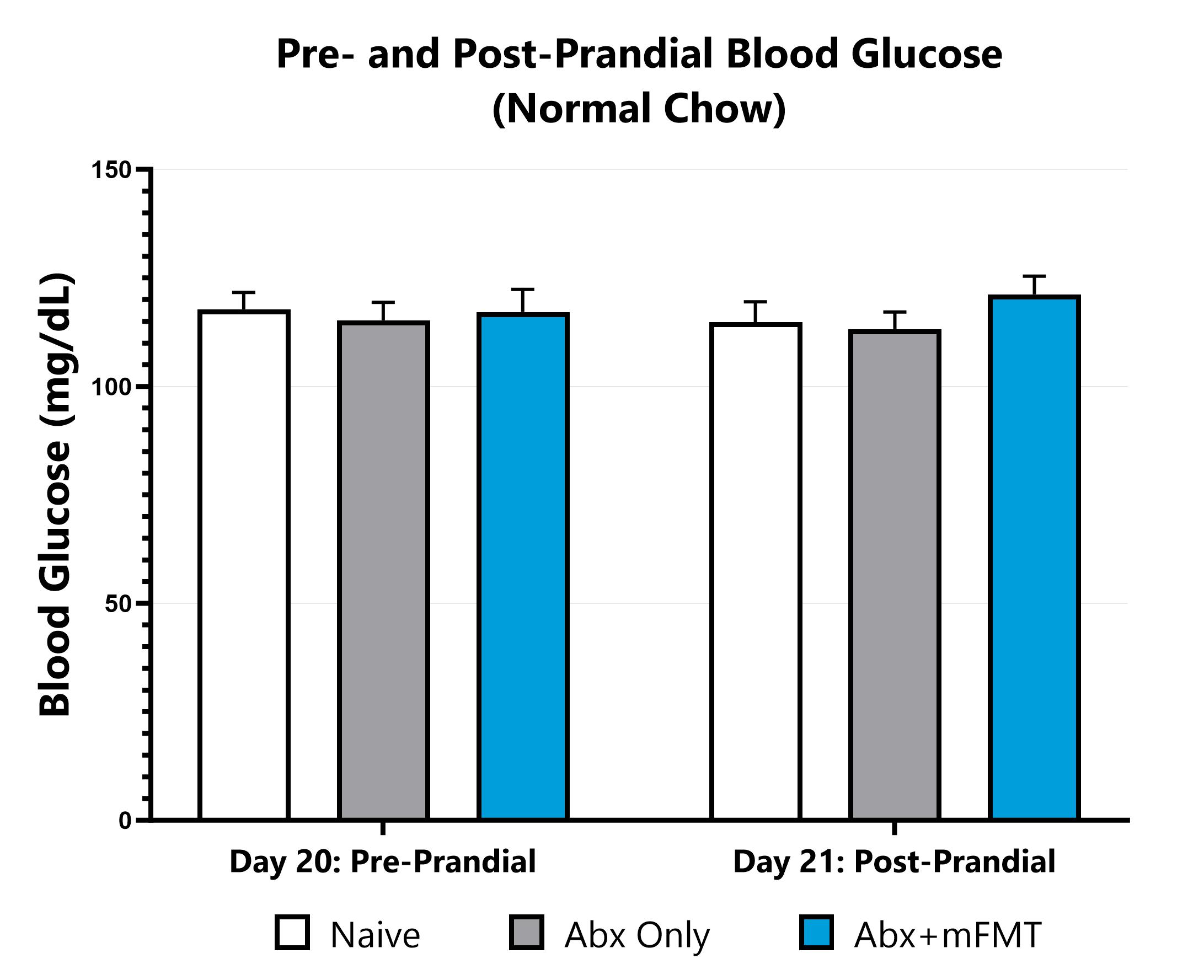
Animals are fasted and blood glucose is measured via glucose monitor. Food is returned to animals and blood glucose is re-measured via glucose monitor the next day.
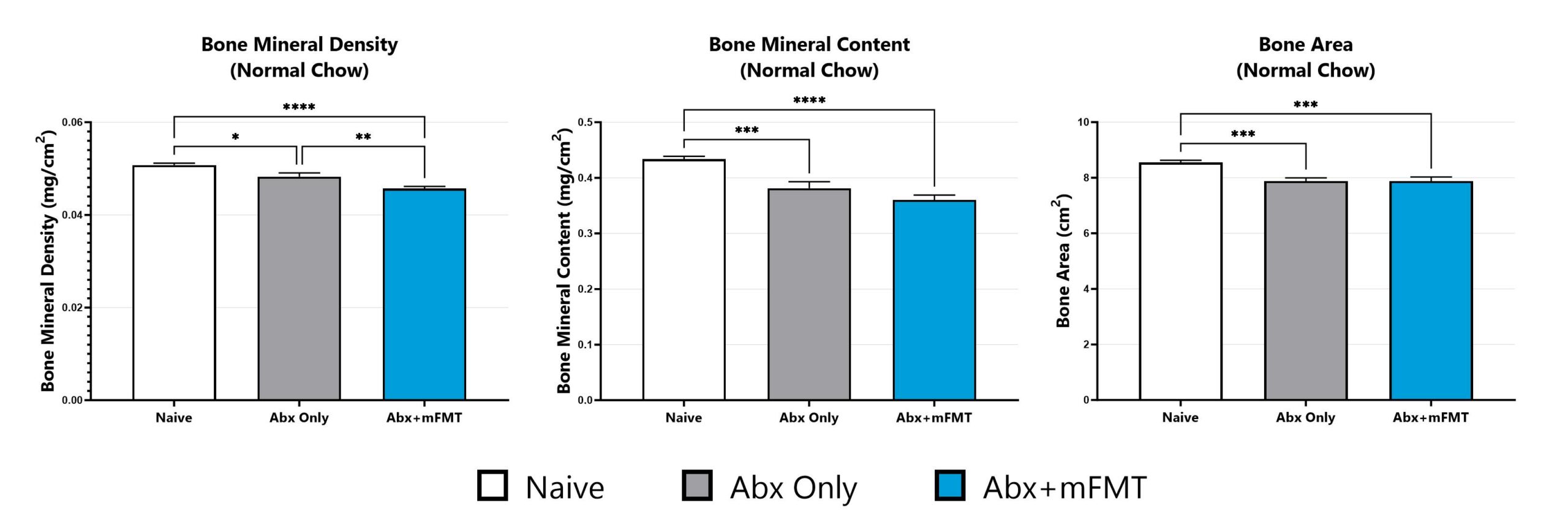
Bone phenotypes are assessed via dual-energy x-ray absorptiometry (DEXA) analysis. (*p<0.05; **p<0.01; ***p<0.005; ****p<0.001).
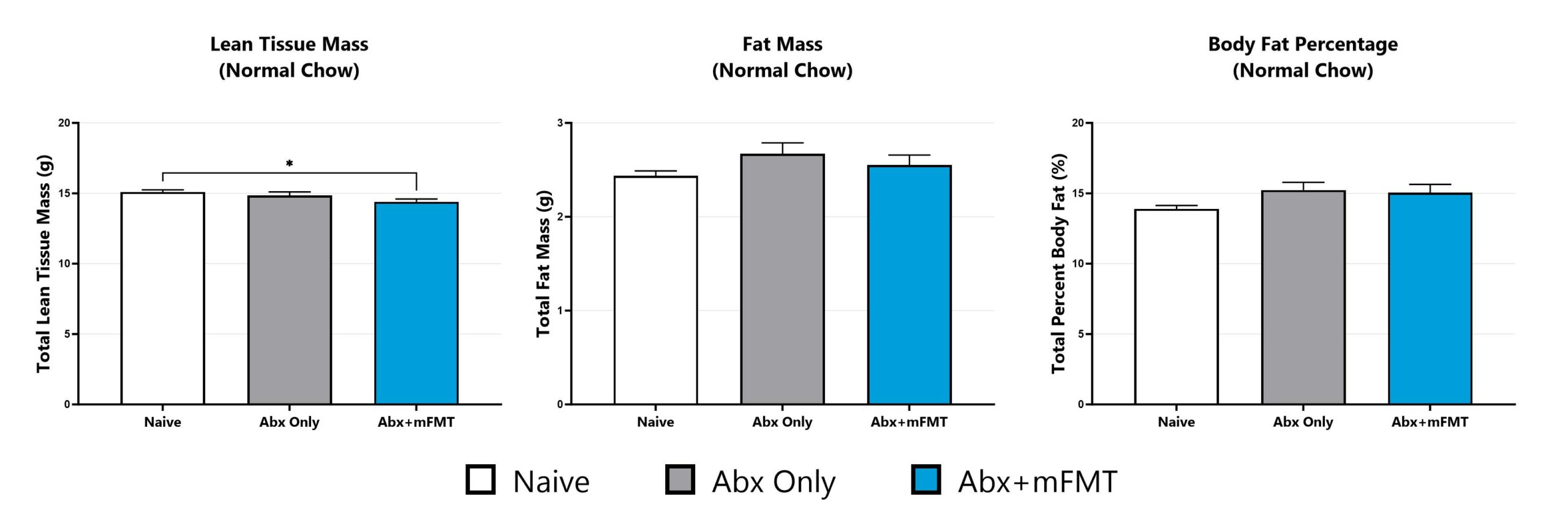
Tissue phenotypes are assessed via dual-energy x-ray absorptiometry (DEXA) analysis. (*p<0.05).
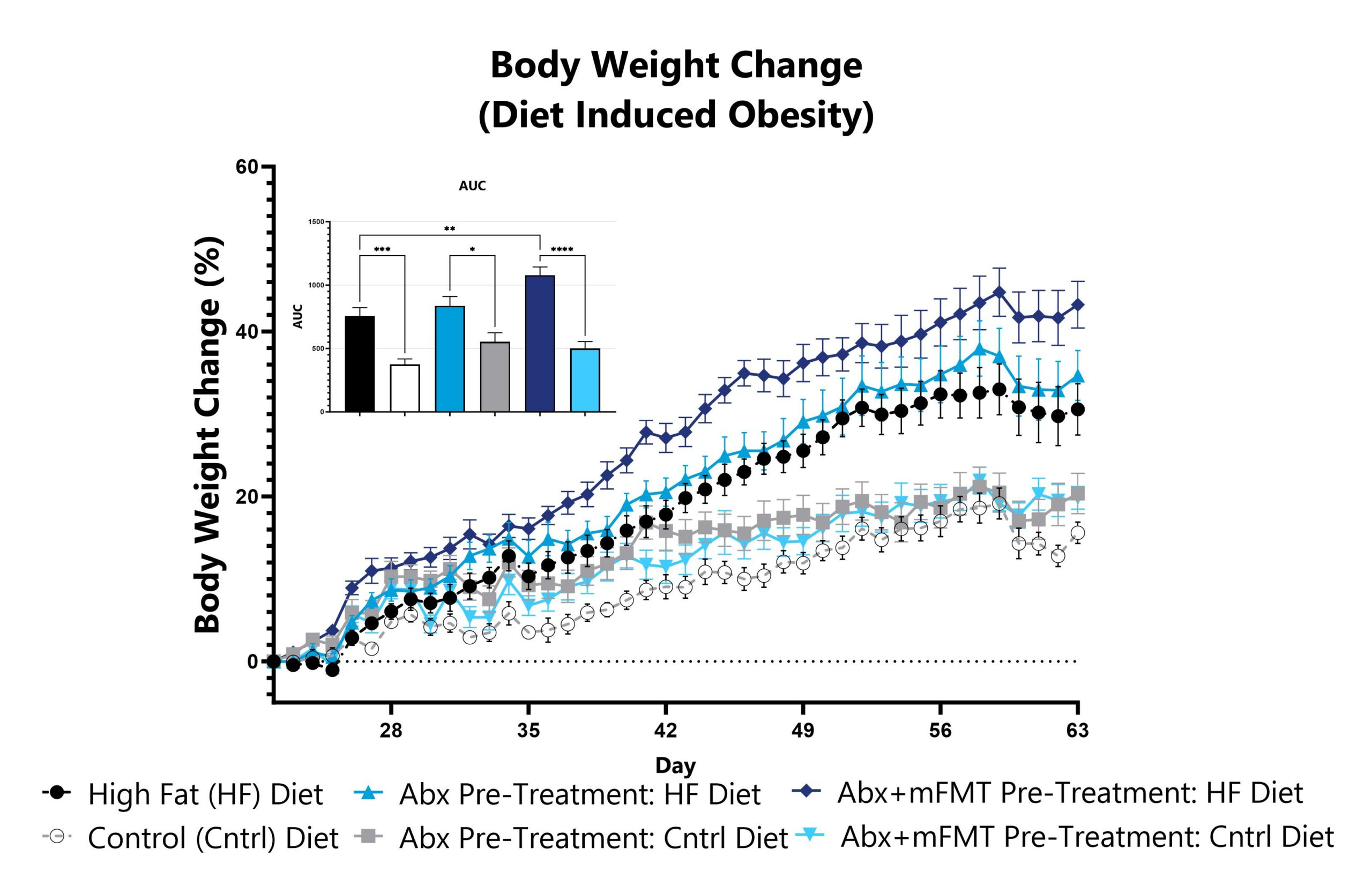
Animals are weighed daily, and body weight change as compared to day of HFD administration (Day 22) is calculated. The AUC is calculated to compare treatment arms and is shown in the inset. (*p<0.05; **p<0.01; ***p<0.005; ****p<0.001).
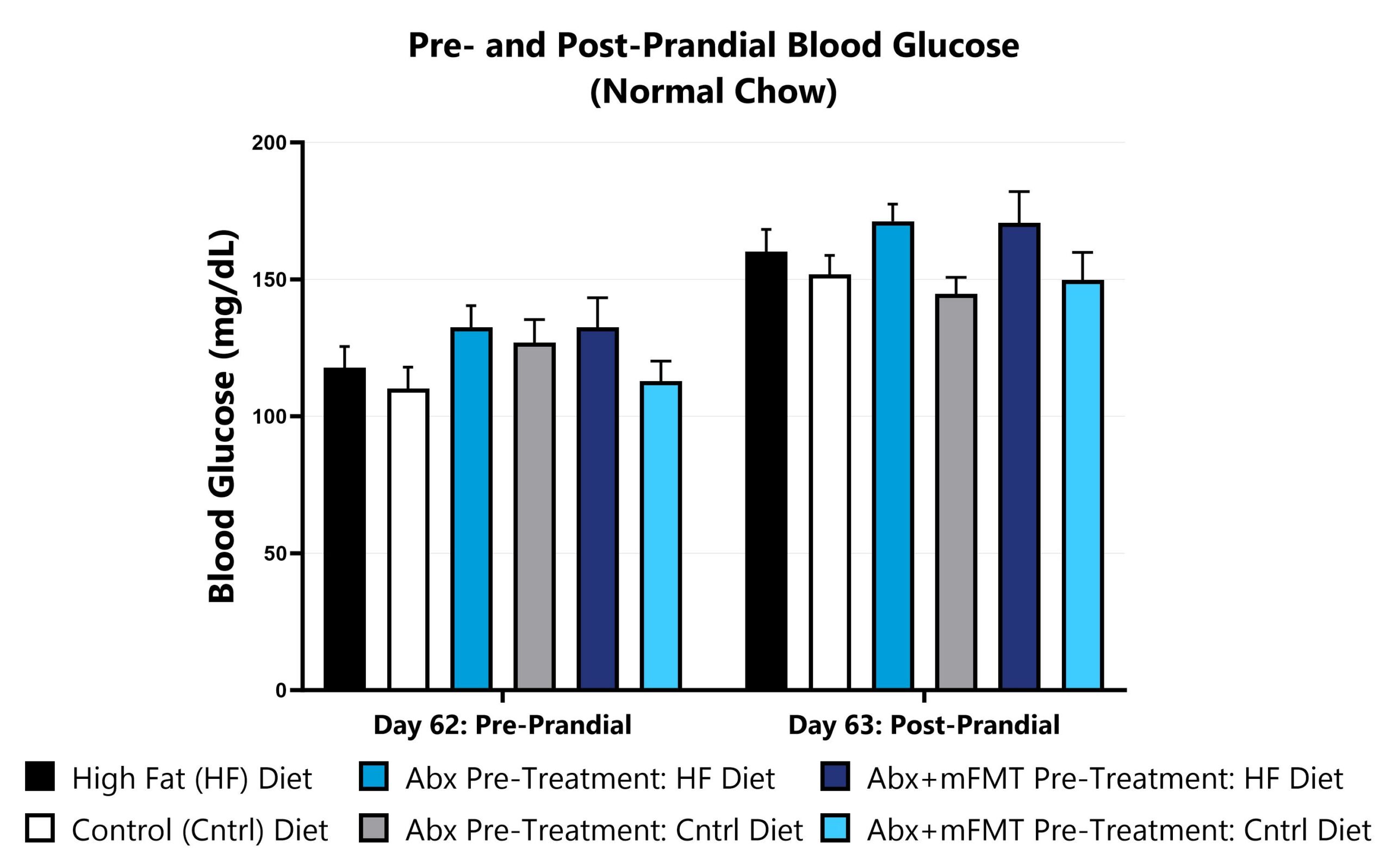
Animals are fasted and blood glucose is measured via glucose monitor. Food is returned to animals and blood glucose is re-measured via glucose monitor the next day.
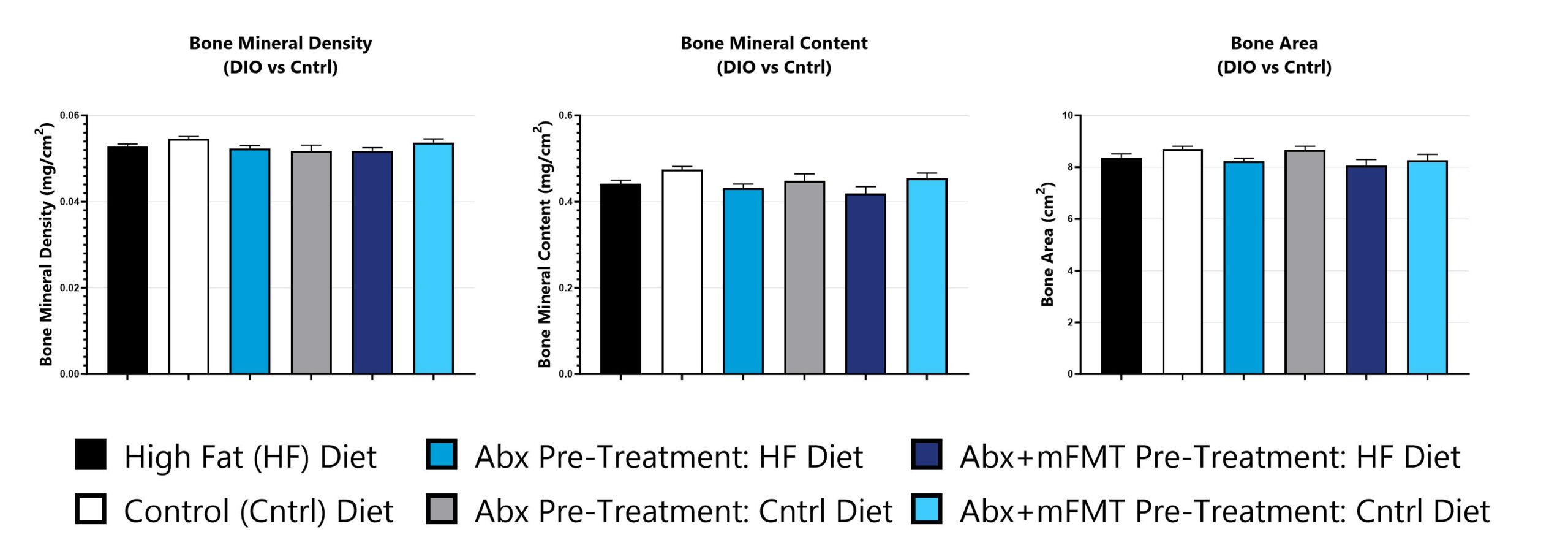
Bone phenotypes are assessed via dual-energy x-ray absorptiometry (DEXA) analysis.
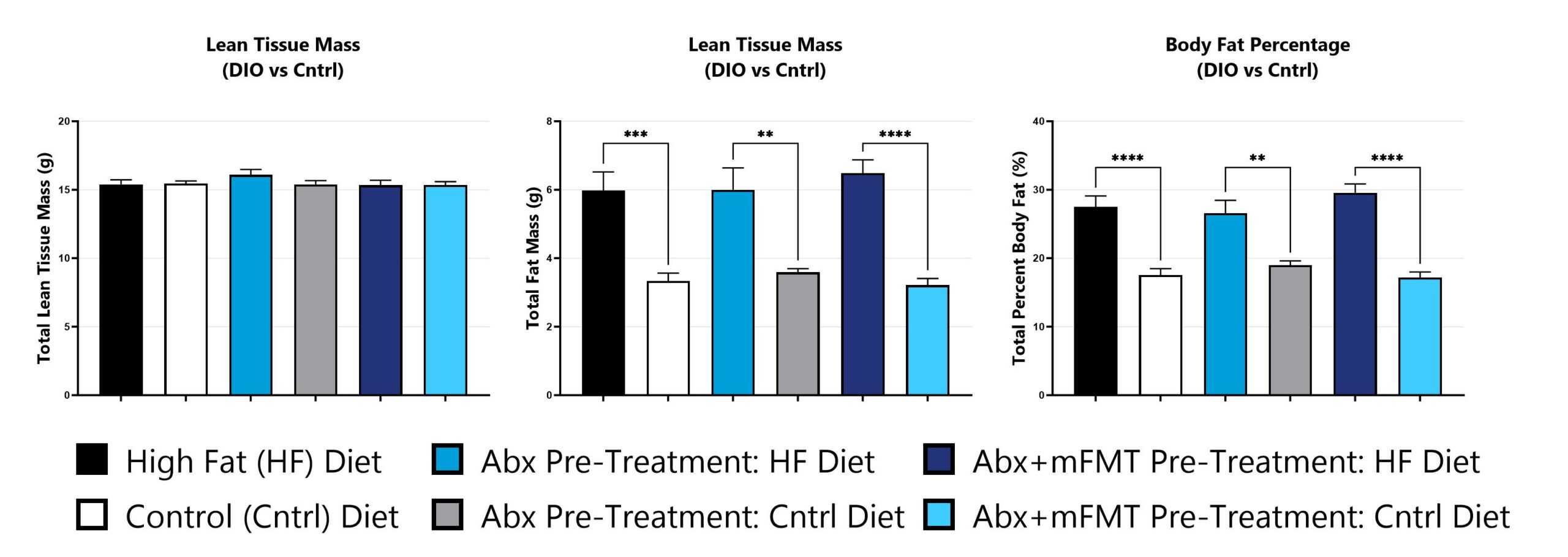
Tissue phenotypes are assessed via dual-energy x-ray absorptiometry (DEXA) analysis. (**p<0.01; ***p<0.005; ****p<0.001).

Close

Differential response to arthritis induction can be observed in animals sourced from different vendors, some of which are mediated by vendor-differences in microbiome. Evaluation of Rheumatoid Arthritis (RA) phenotypes in animals sourced from different vendors can be a straightforward approach to assess the microbiome’s impact in a given model. Data from the Glucose-6-phosphate isomerase (GPI)-Induced Arthritis Model induced in animals from different vendors is shown as an example.
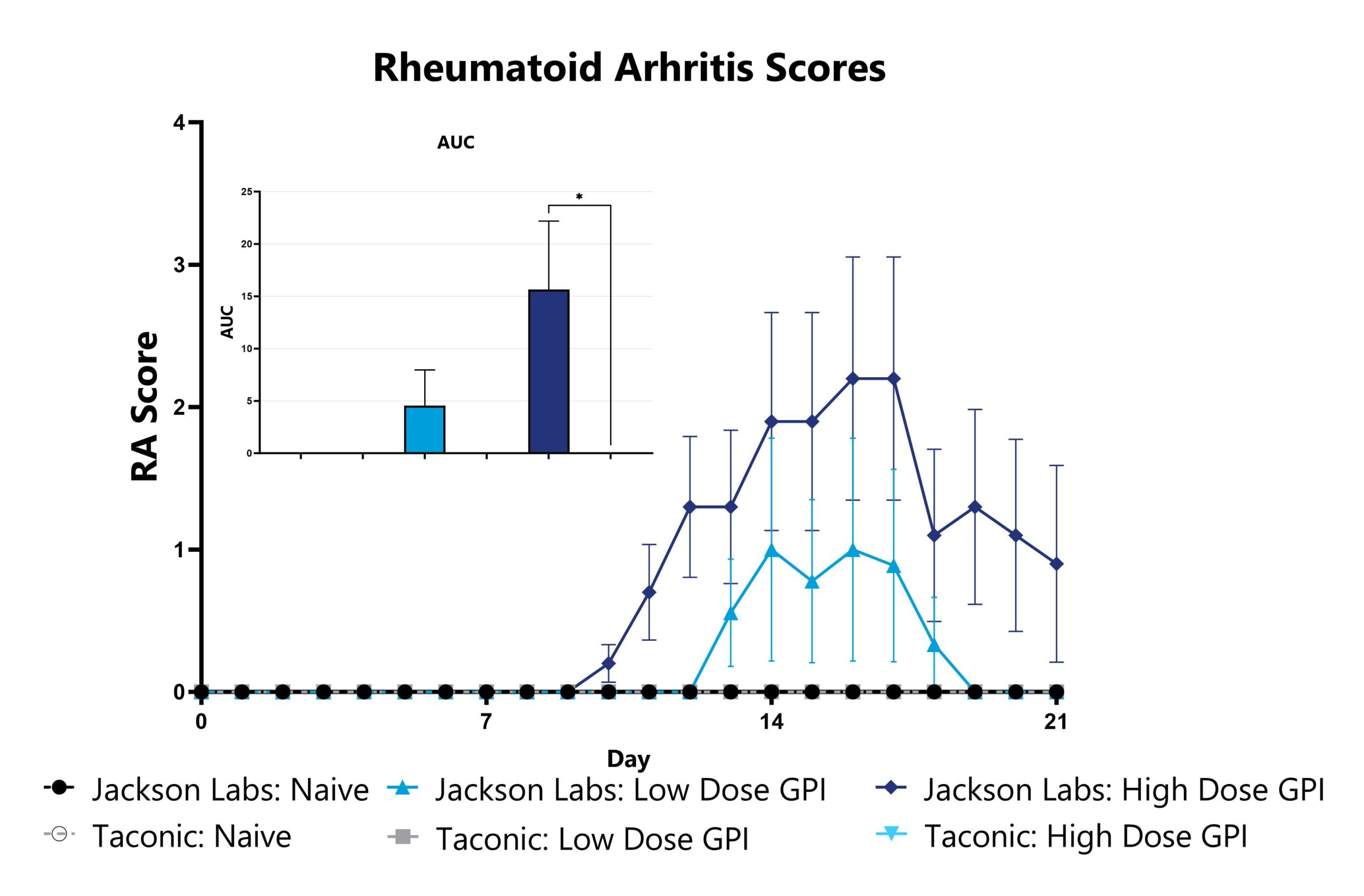
Animals are monitored for signs of arthritis on a daily basis and assigned RA scores. The AUC is calculated to compare treatment arms and is shown in the inset. (*p<0.05).
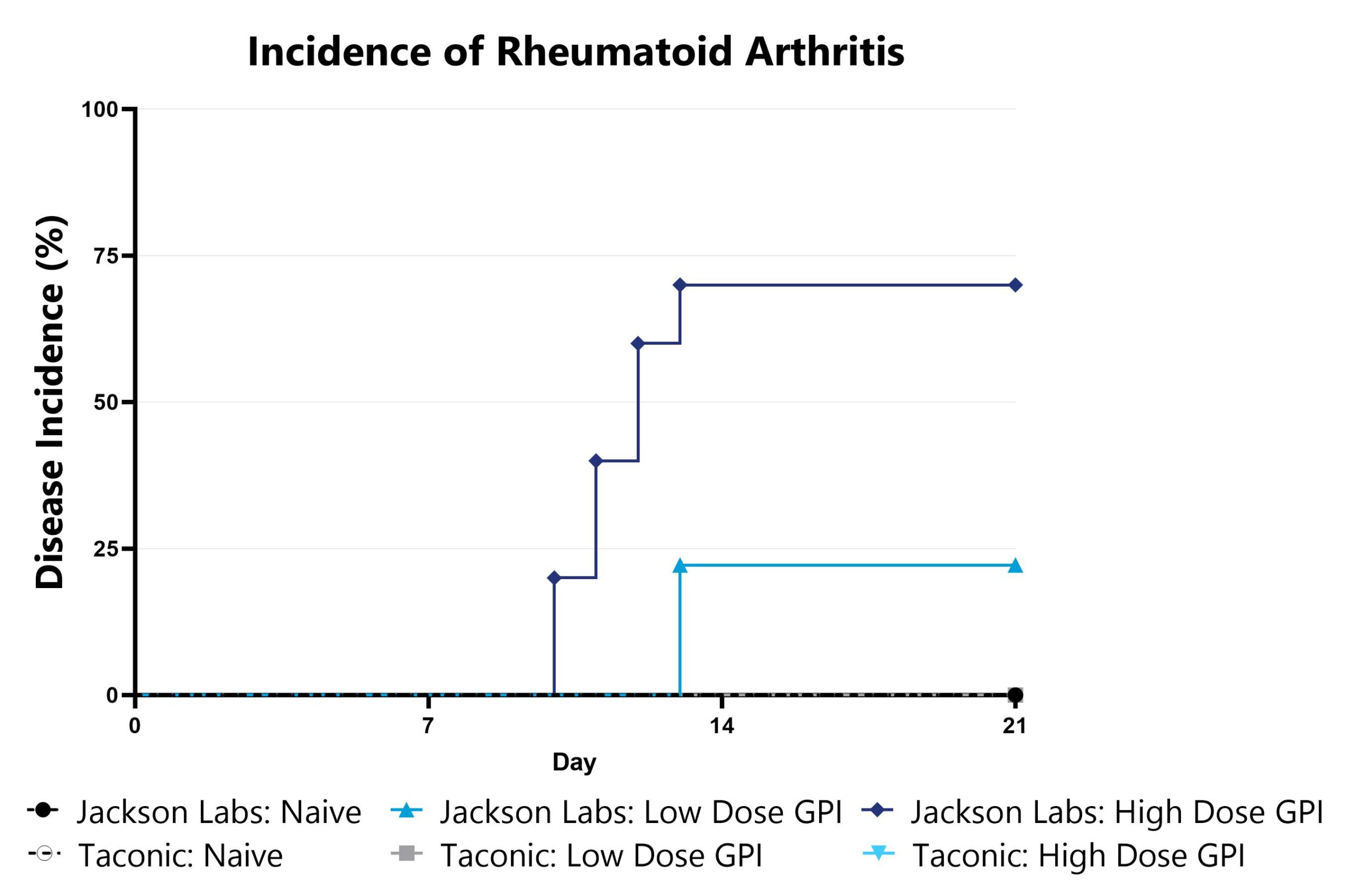
Animals are monitored for signs of arthritis on a daily basis and overall disease incidence is shown.

Close



