- Research Services
- Capabilities
- About Us
- Resources
- Contact Us
 Immunogenicity Evaluation
Immunogenicity Evaluation Infectious Disease Models
Infectious Disease Models Immunogenicity and Infectious Disease
Immunogenicity and Infectious DiseaseDespite consistent advancements in medicine and improvements in general hygiene practices, infectious diseases remain a significant global health burden with disproportionate impact to vulnerable populations. Animal models of infectious disease are widely used to assess the safety and efficacy of vaccine candidates and novel treatments.
BioModels has the capabilities and expertise to evaluate the immunogenicity of established and experimental materials in naïve animals and in a variety of infectious disease models.
Characterizing the immunogenic properties of potential vaccines and therapeutics is an essential step in establishing a complete safety and efficacy profile for the material of interest.
BioModels has the experience and flexibility to develop and execute client-specific programs to characterize both prophylactic and therapeutic modalities with diverse endpoint assessments including but not limited to:
Given the persistent global burden of infectious diseases, there is a clear and acute need for continued research and development for novel intervention methods targeting viral, bacterial, fungal, and parasitic infections.
BioModels has experience with a variety of these models and a fully equipped BSL-2 facility to support pre-clinical programs with the evaluation of customizable, experimental endpoints such as:
Study Models

Type IV Delayed-type hypersensitivity (DTH) is a cell-mediated, antigen-specific immune reaction, generally predictive of T cell function and response. The Type IV DTH model is an important clinical tool that has been widely adopted and can be useful for allergy testing, determining immune resistance to pathogens, predicting vaccine immunogenicity, and more. This model typically includes a sensitization phase, where the animals will be administered a test article that will expose them to the antigen of interest. The animals will then be subject to a challenge with the same antigen by an intradermal ear injection. The T cell activation and resulting inflammation can be assessed with physical measurements of the localized swelling and downstream blood and tissue analyses as outlined below.

Close

Pre-clinical assessment of the immunogenicity of potential vaccine or therapeutic candidates can provide valuable insight for safety and efficacy profiles. BioModels can provide custom designed programs to characterize antigen-specific immune responses. Representative data depicts an example assessment of ex vivo T cell responses from naïve mice following PMA/Ionomycin stimulation. Cytokine panels and stimulation paradigms are fully customizable.
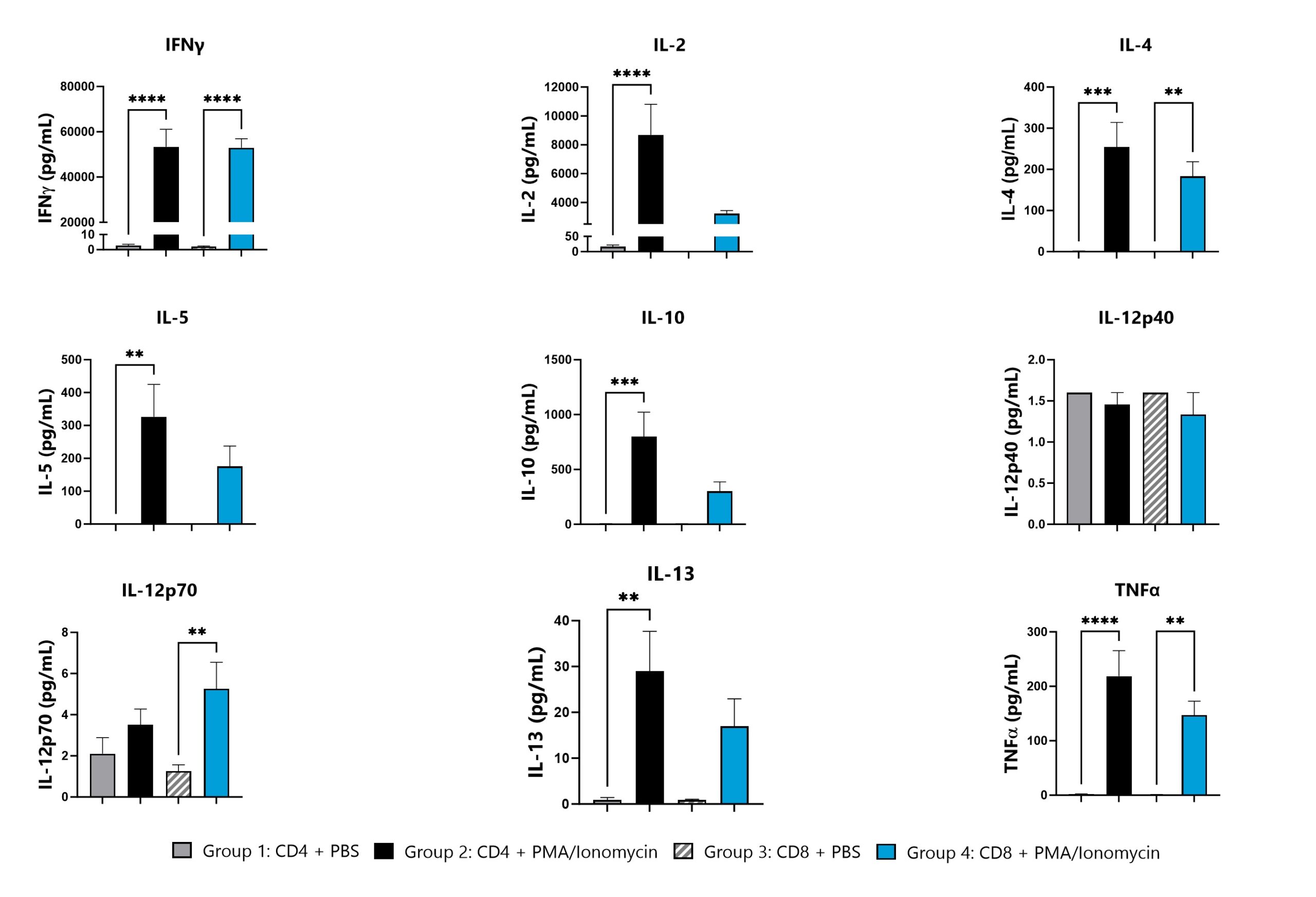
Spleen and iLN cells harvested from naive animals are stimulated with PBS or PMA/Ionomycin. Cell culture supernatants are then assessed for cytokine levels. Stimulated groups are compared to their respective vehicle control group (**p<0.01; ***p<0.001; ****p<0.0001).

Close
Study Models

Citrobacter rodentium is a murine, gram-negative, facultative anaerobic enteropathogen with genetic properties and virulence factors that evoke an inflammatory response similar to that induced in humans by enteropathogenic Escherichia coli (EPEC). Infectious colitis induced with C. rodentium is therefore a clinically relevant, translational model of intestinal dysbiosis with a range of phenotypic severity based on the host mouse strain. This model can be utilized to assess the efficacy of novel therapeutics against EPEC or to characterize the behavior of test compounds in a pathogen-induced, inflammatory setting. C. rodentium infection in C57Bl/6 mice via oral gavage induces self-limiting disease that is typically characterized by weight loss, increased rates of diarrhea/blood in the stool, and colonic inflammation. The representative data depicts a model of C. rodentium infectious colitis with a microbiome targeted treatment paradigm of fecal microbiota transfers from animals with segmented filamentous bacteria (SFB) into confirmed SFB- animals.
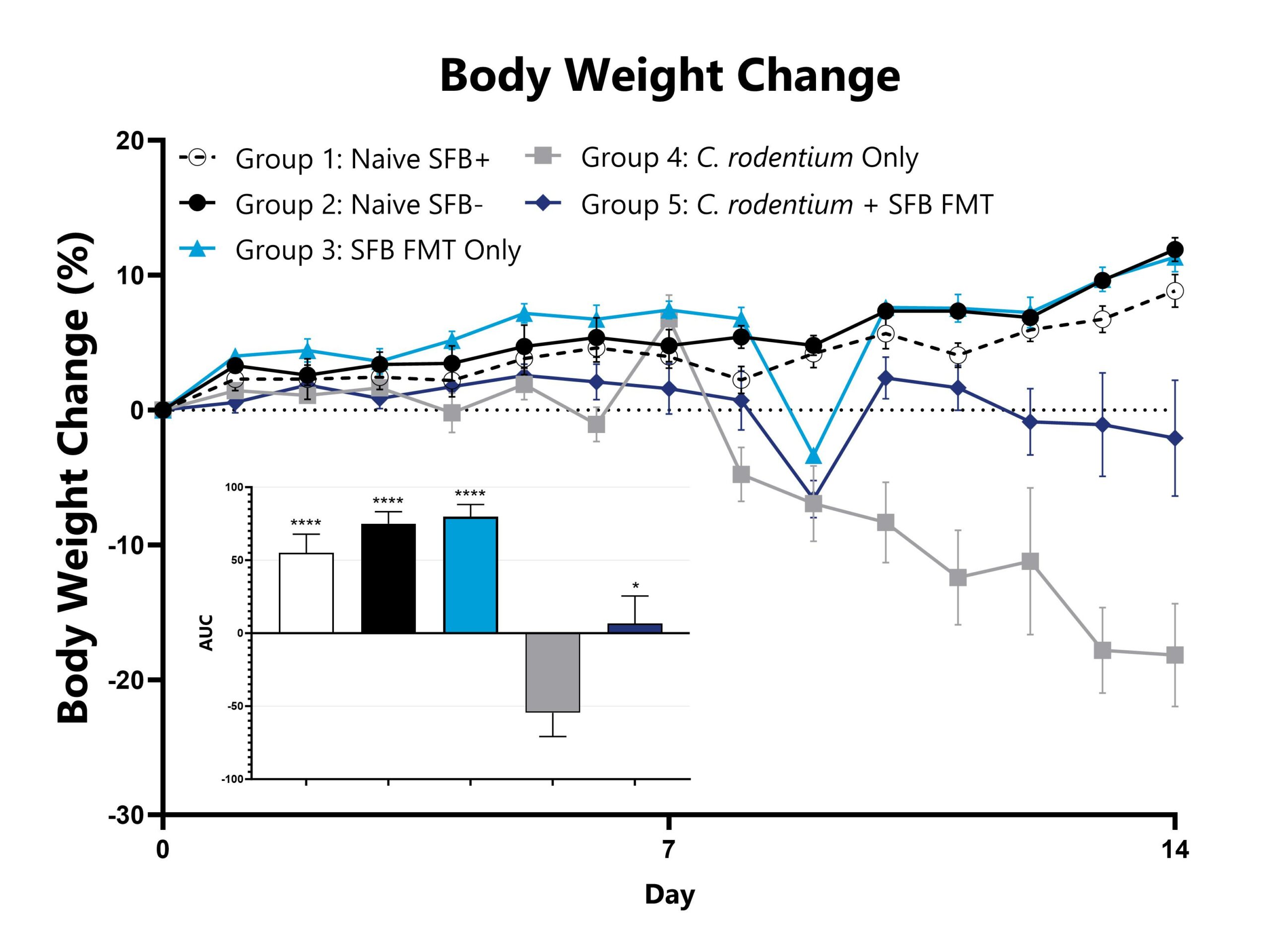
Animals are weighed daily, and percent body weight change relative to Day 0 is calculated. AUCs are calculated to compare groups and are shown in the figure inset (*p<0.05; ****p<0.0001).
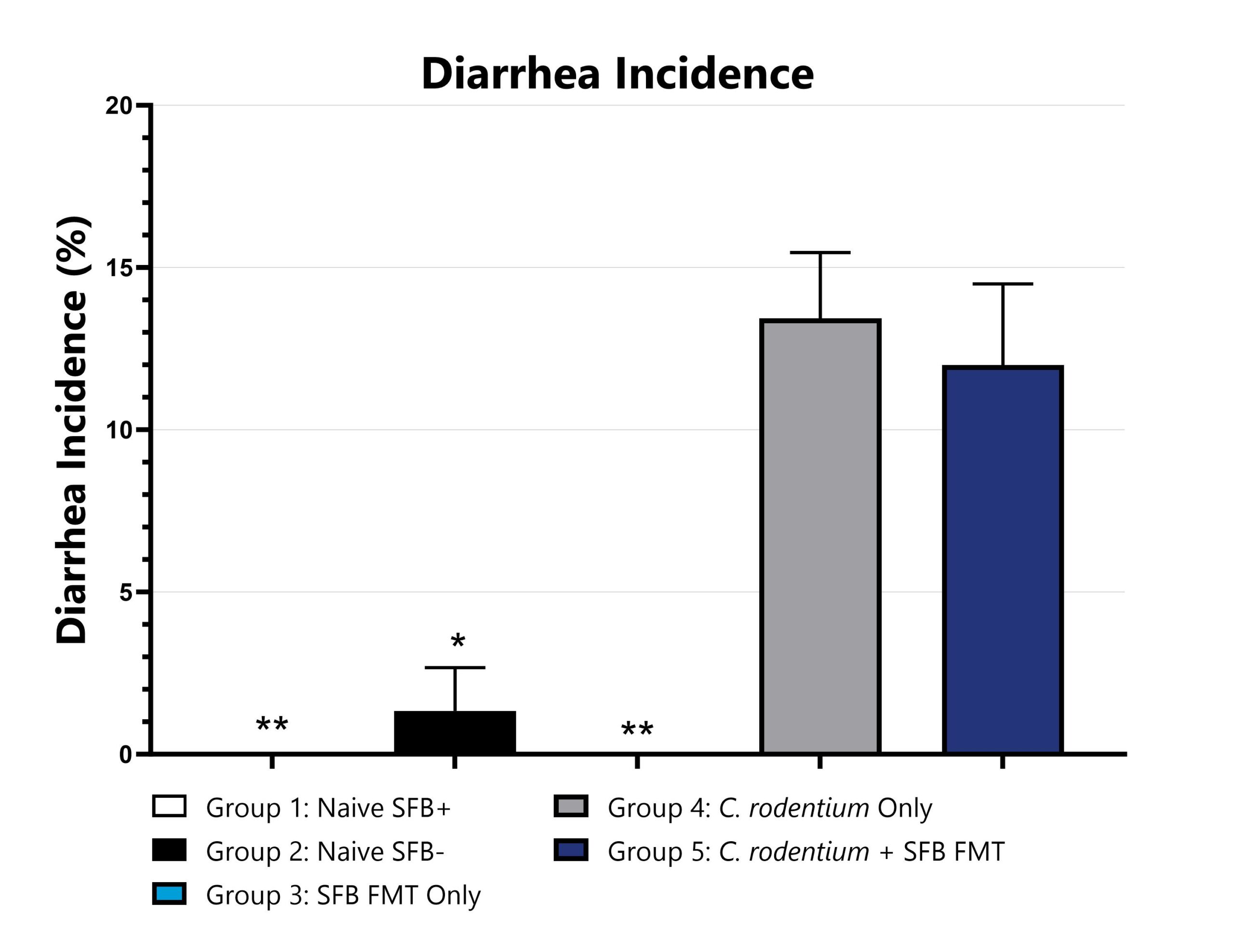
Animals are observed for diarrhea on a daily basis and the overall incidence rates are calculated and compared to Group 4 (*p<0.05; **p<0.01).
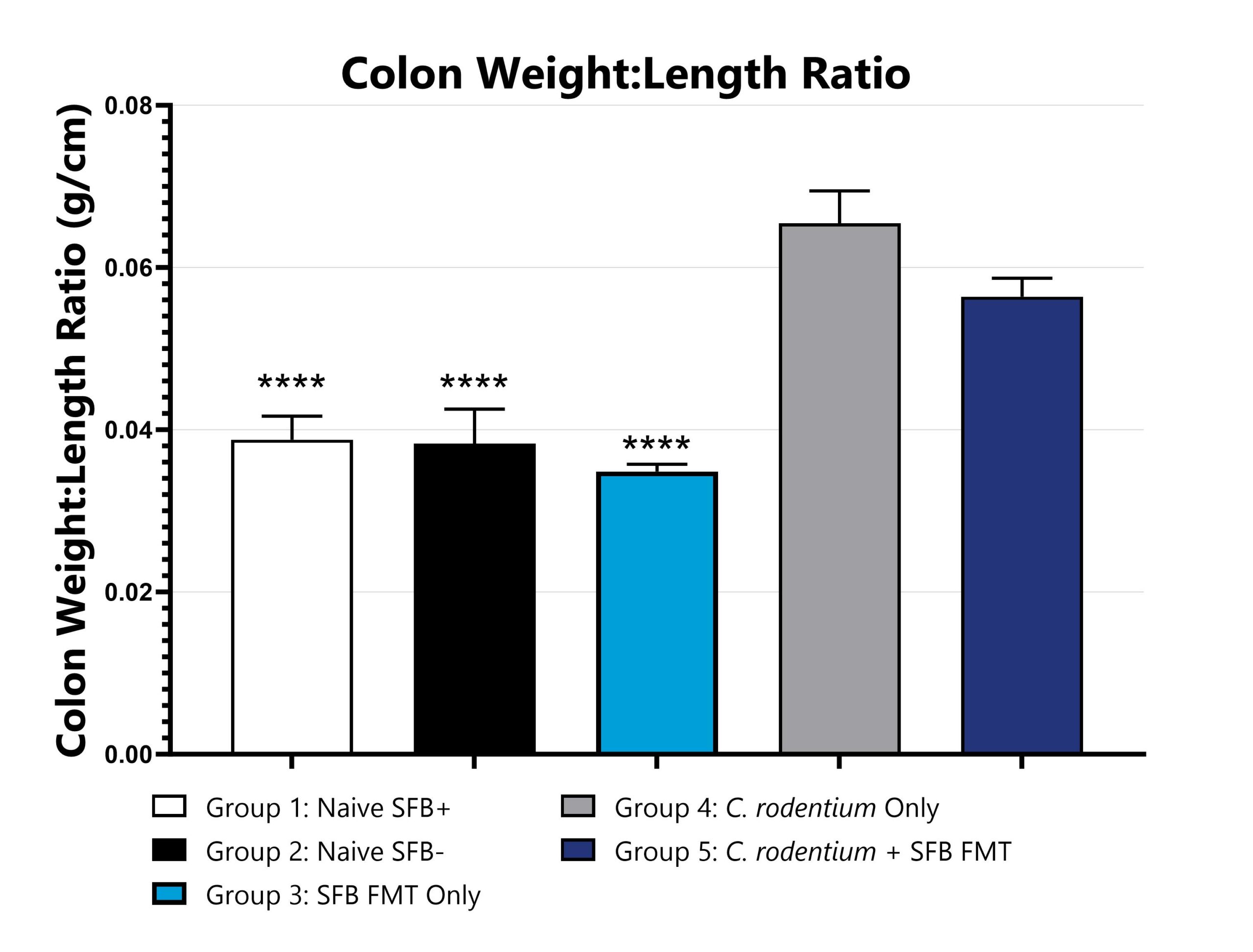
Colons are excised, measured, and weighed on Day 14. Colon weight:length ratios are calculated and compared to Group 4 (****p<0.0001).
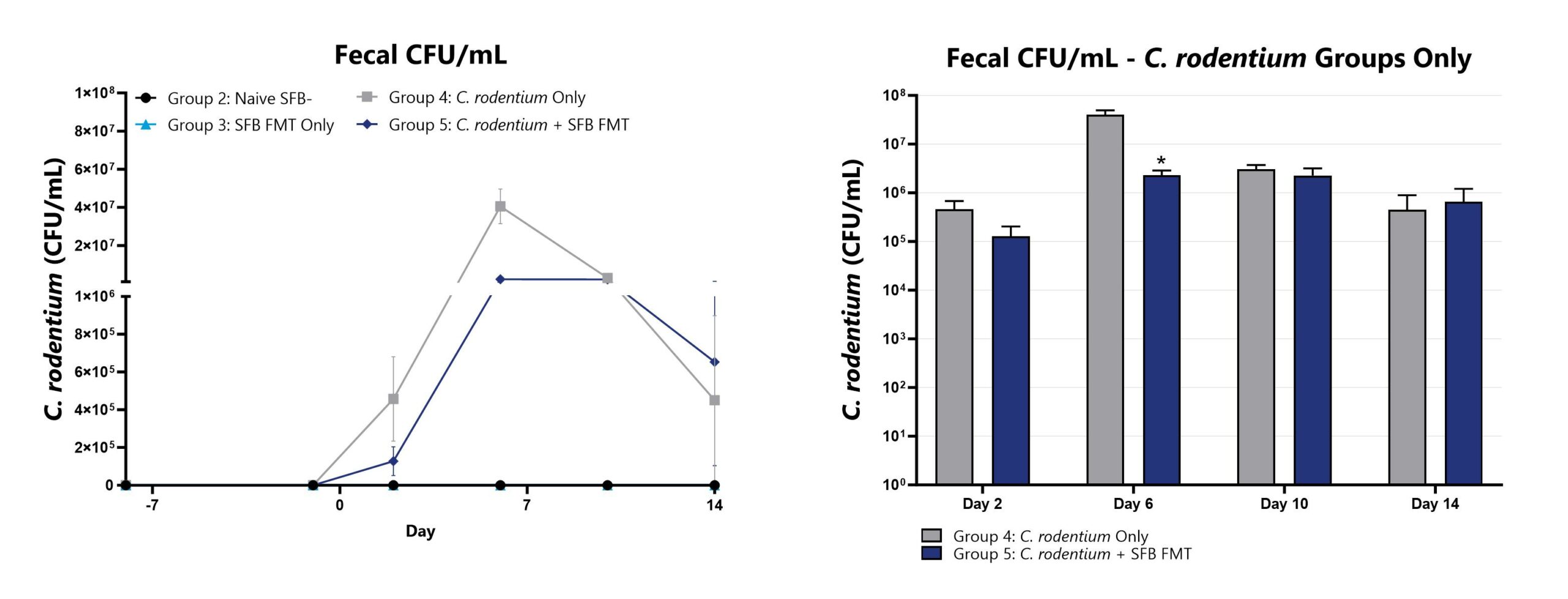
Feces are collected throughout the study from Groups 2-5, homogenized, and plated for C. rodentium CFU enumeration. All groups are compared to Group 4 (*p<0.05; **p<0.01).
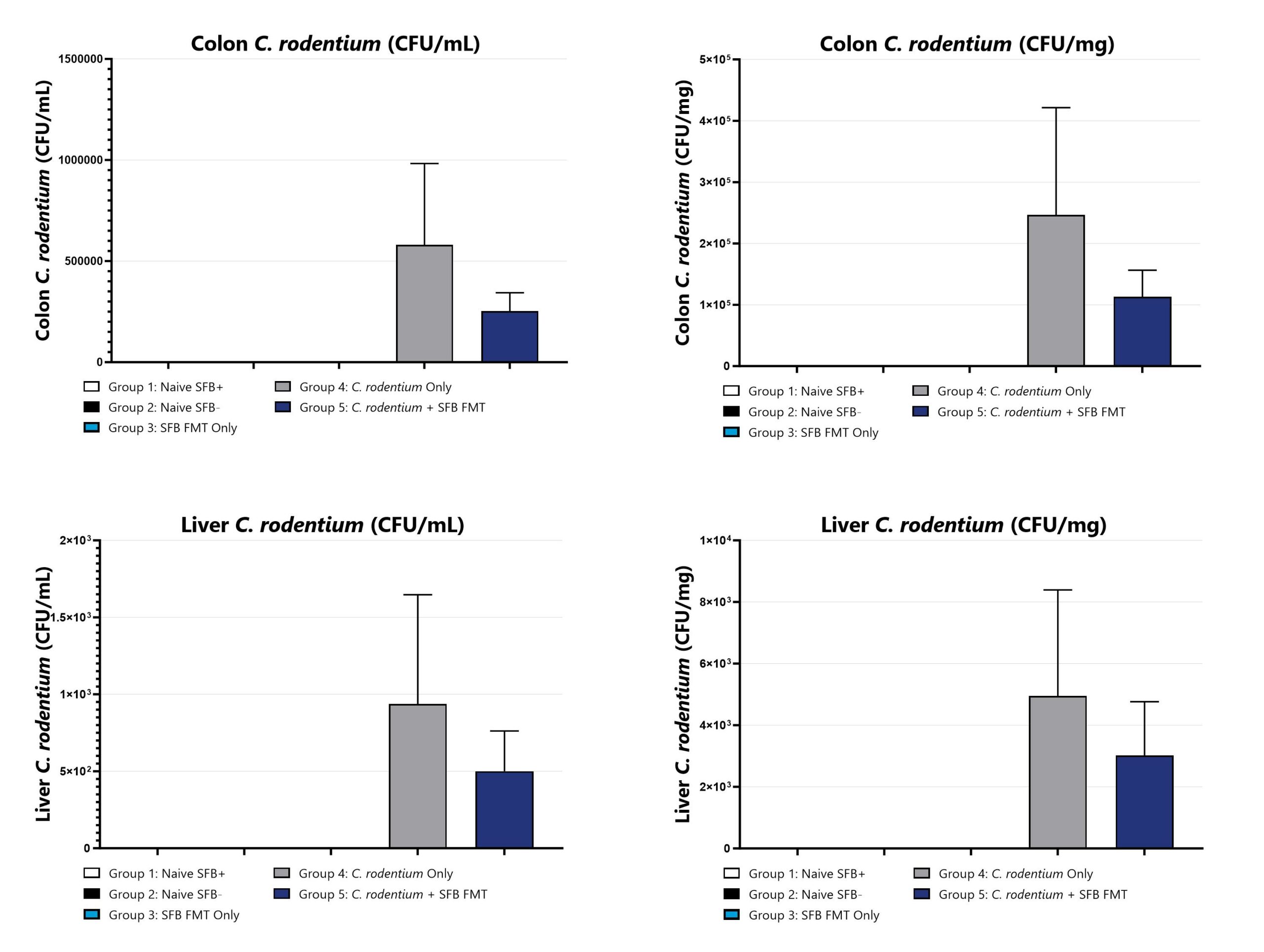
Day 14 livers and cecums/colons are collected from all groups, homogenized, and plated for C. rodentium CFU enumeration. CFU/mL (left) and CFU/g (right) are shown.

Close

Salmonella enterica is a gram-negative, facultative anaerobic pathogen that can impact the gastrointestinal systems of both human and animal populations. S. enterica subspecies 1 serovar Typhimurium is the primary etiological agent of foodborne salmonellosis resulting in non-systemic enterocolitis in humans. Oral infection of susceptible C57Bl/6 mice with serovar Typhimurium is an established in vivo model used to study the pathogenesis of Salmonella-induced gastroenteritis and to assess the efficacy of novel therapeutics. This infection is characterized by weight loss, epithelial barrier disruption, and increased inflammation in the gastrointestinal tract. Representative data demonstrates kinetics of disease severity over 72 hours with body weight tracking, assessment of colonic infiltrating neutrophils with correlating MPO production, and histopathology analysis of colon segments.

Animals are weighed daily, and percent body weight change relative to Day 0 is calculated. AUCs are calculated to compare infected and uninfected animals and are shown in the figure inset (***p<0.001).
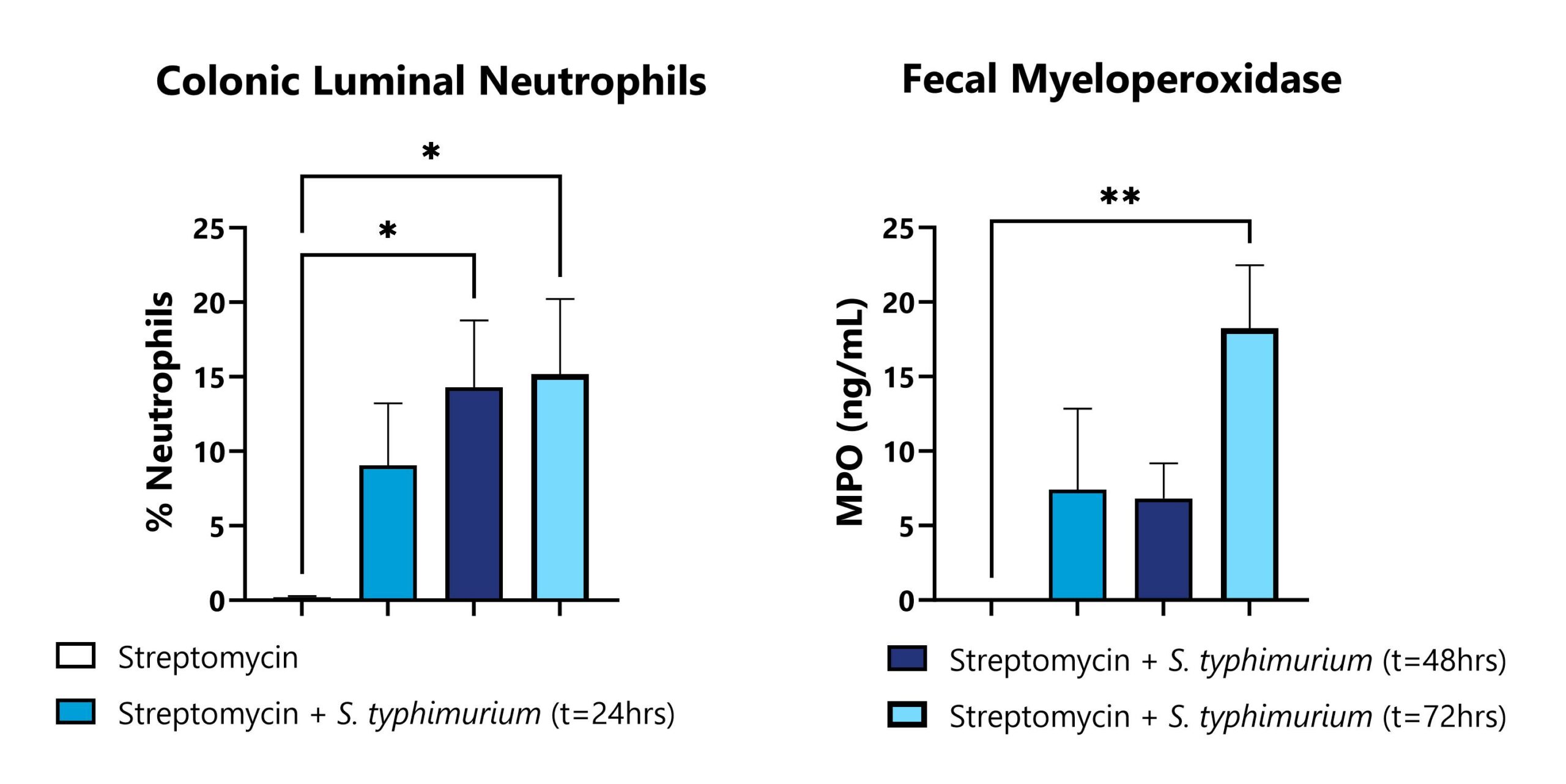
Colons and fecal pellets are collected from infected animals. Colon tissue is processed and analyzed via FACS for neutrophil populations. Fecal homogenates are assessed for myeloperoxidase levels. Infected groups were compared to naïve animals (*p<0.05; **p<0.01).

Colons of infected animals at t=24, 48, and 72 hours are compared with naïve colons via histopathology analysis (*p<0.05; **p<0.01).
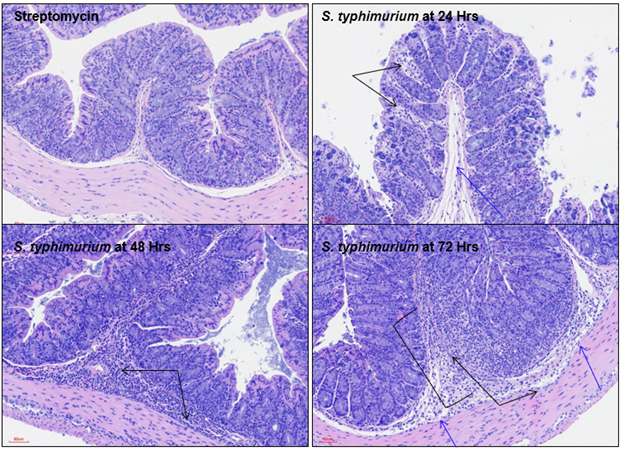
Representative H&E stained colon samples from infected animals at t=24, 48, and 72hrs compared to a naïve colon. Black arrows denote inflammation. Blue arrows denote edema. Black brackets denote mucosal necrosis. Histopathology performed by Dallas Tissue Research.

Close

Candida albicans is the most commonly identified pathogenic Candida species in human infections and can impact both immunocompromised and healthy populations. C. albicans has adapted an expansive set of virulence factors and fitness attributes that enable this fungus to survive in extremely diverse environments and infect humans and animals through a variety of routes. This model could be used to assess candidate anti-fungal treatment efficacy or characterize the biodistribution of test articles in a model of systemic inflammation due to infection. Representative data shows infection kinetics including body weight, survival, and CFU in critical organs for multiple different routes of infection with C. albicans.
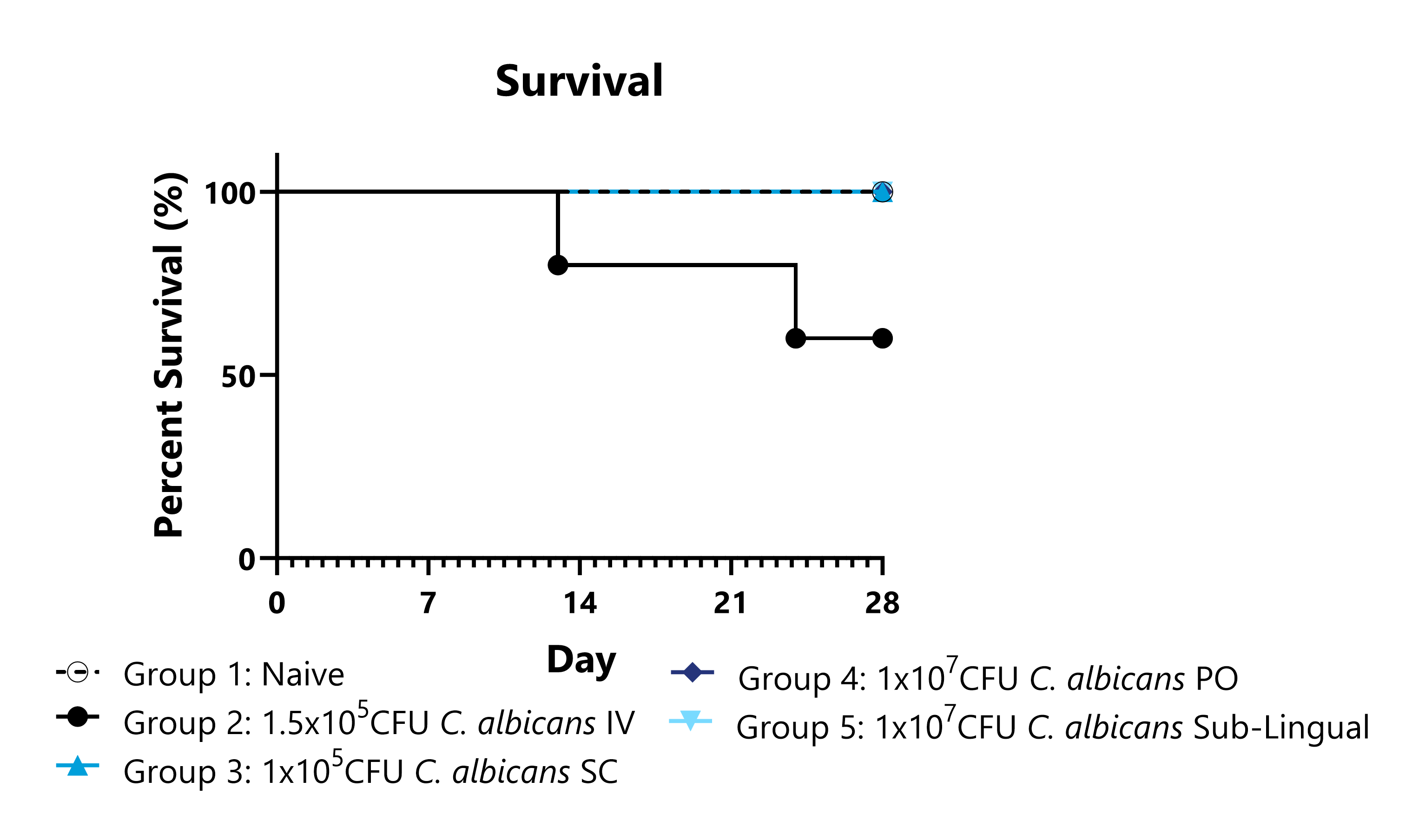
Animals are observed daily for survival and morbidity. All infected groups are compared to naïve animals to assess for statistical significance.
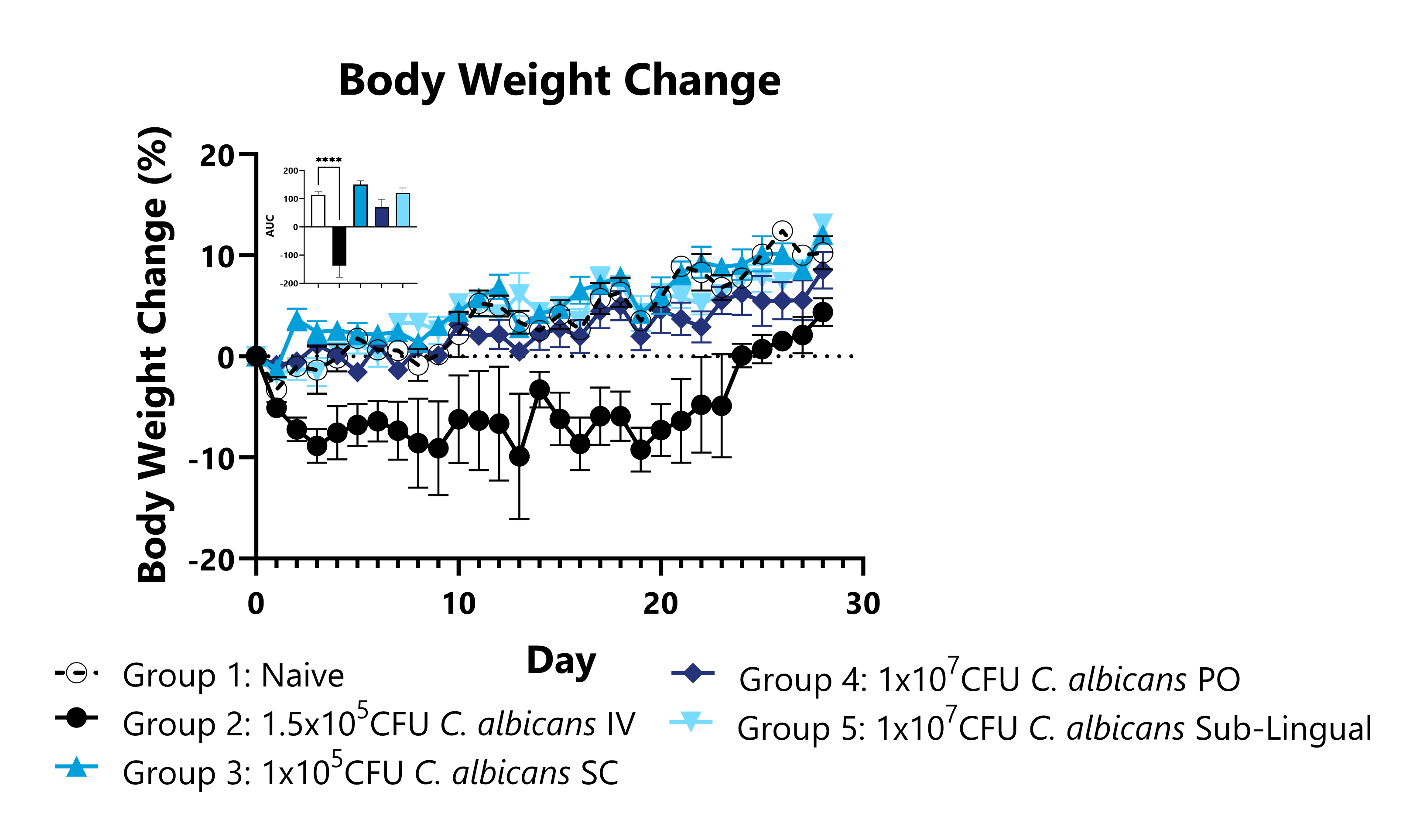
Animals are weighed daily, and percent body weight change relative to Day 0 is calculated. AUCs are calculated to compare infected and uninfected animals and are shown in the figure inset (****p<0.0001).
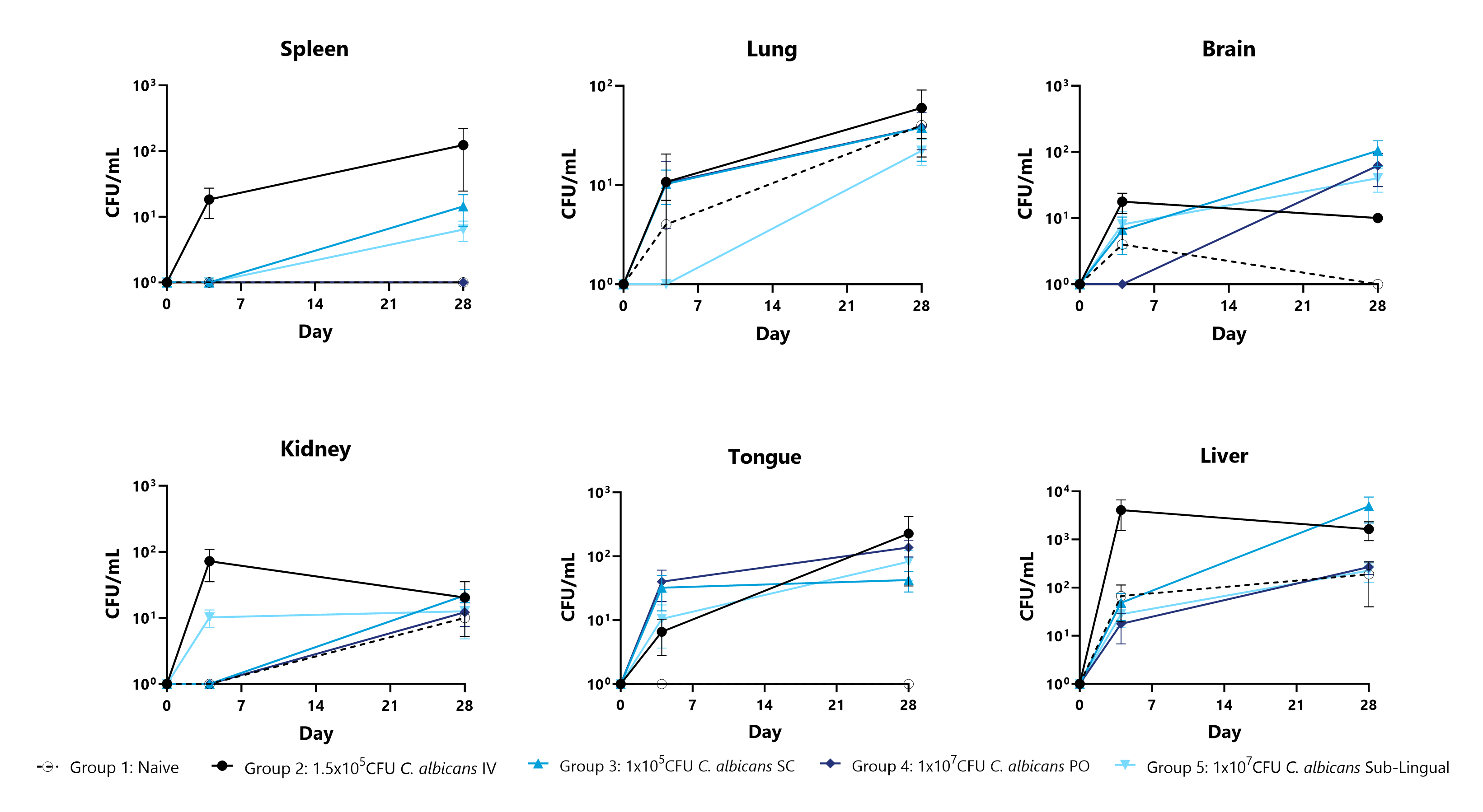
Animals are euthanized 4 and 28 days post infection. Spleens, lungs, brains, kidneys, tongues, and livers are excised, processed, and plated for CFU enumeration.

Close



